Andrew Laine
Push the Boundary of SAM: A Pseudo-label Correction Framework for Medical Segmentation
Aug 02, 2023Abstract:Segment anything model (SAM) has emerged as the leading approach for zero-shot learning in segmentation, offering the advantage of avoiding pixel-wise annotation. It is particularly appealing in medical image segmentation where annotation is laborious and expertise-demanding. However, the direct application of SAM often yields inferior results compared to conventional fully supervised segmentation networks. While using SAM generated pseudo label could also benefit the training of fully supervised segmentation, the performance is limited by the quality of pseudo labels. In this paper, we propose a novel label corruption to push the boundary of SAM-based segmentation. Our model utilizes a novel noise detection module to distinguish between noisy labels from clean labels. This enables us to correct the noisy labels using an uncertainty-based self-correction module, thereby enriching the clean training set. Finally, we retrain the network with updated labels to optimize its weights for future predictions. One key advantage of our model is its ability to train deep networks using SAM-generated pseudo labels without relying on a subset of expert-level annotations. We demonstrate the effectiveness of our proposed model on both X-ray and lung CT datasets, indicating its ability to improve segmentation accuracy and outperform baseline methods in label correction.
Cardiac Adipose Tissue Segmentation via Image-Level Annotations
Jun 09, 2022



Abstract:Automatically identifying the structural substrates underlying cardiac abnormalities can potentially provide real-time guidance for interventional procedures. With the knowledge of cardiac tissue substrates, the treatment of complex arrhythmias such as atrial fibrillation and ventricular tachycardia can be further optimized by detecting arrhythmia substrates to target for treatment (i.e., adipose) and identifying critical structures to avoid. Optical coherence tomography (OCT) is a real-time imaging modality that aids in addressing this need. Existing approaches for cardiac image analysis mainly rely on fully supervised learning techniques, which suffer from the drawback of workload on labor-intensive annotation process of pixel-wise labeling. To lessen the need for pixel-wise labeling, we develop a two-stage deep learning framework for cardiac adipose tissue segmentation using image-level annotations on OCT images of human cardiac substrates. In particular, we integrate class activation mapping with superpixel segmentation to solve the sparse tissue seed challenge raised in cardiac tissue segmentation. Our study bridges the gap between the demand on automatic tissue analysis and the lack of high-quality pixel-wise annotations. To the best of our knowledge, this is the first study that attempts to address cardiac tissue segmentation on OCT images via weakly supervised learning techniques. Within an in-vitro human cardiac OCT dataset, we demonstrate that our weakly supervised approach on image-level annotations achieves comparable performance as fully supervised methods trained on pixel-wise annotations.
Recursive Refinement Network for Deformable Lung Registration between Exhale and Inhale CT Scans
Jun 14, 2021
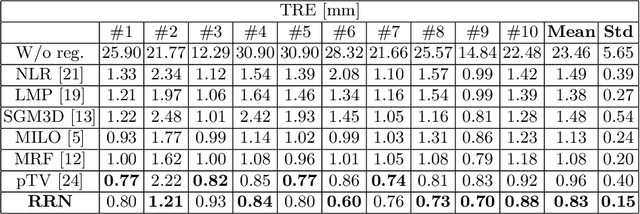
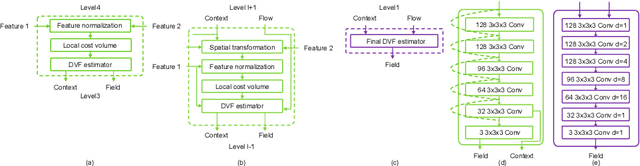
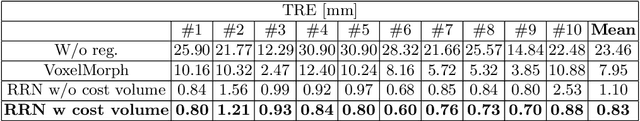
Abstract:Unsupervised learning-based medical image registration approaches have witnessed rapid development in recent years. We propose to revisit a commonly ignored while simple and well-established principle: recursive refinement of deformation vector fields across scales. We introduce a recursive refinement network (RRN) for unsupervised medical image registration, to extract multi-scale features, construct normalized local cost correlation volume and recursively refine volumetric deformation vector fields. RRN achieves state of the art performance for 3D registration of expiratory-inspiratory pairs of CT lung scans. On DirLab COPDGene dataset, RRN returns an average Target Registration Error (TRE) of 0.83 mm, which corresponds to a 13% error reduction from the best result presented in the leaderboard. In addition to comparison with conventional methods, RRN leads to 89% error reduction compared to deep-learning-based peer approaches.
PTNet: A High-Resolution Infant MRI Synthesizer Based on Transformer
May 28, 2021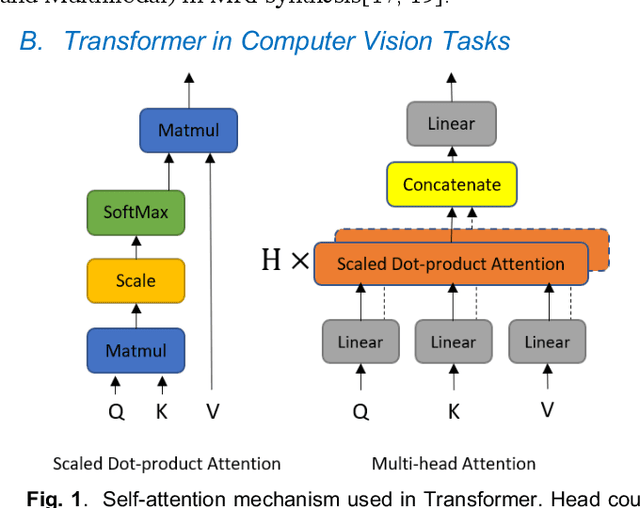
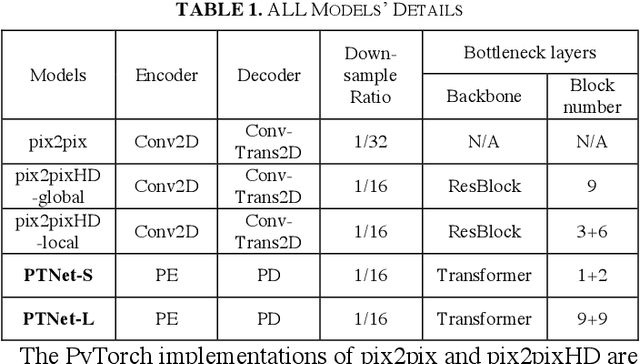
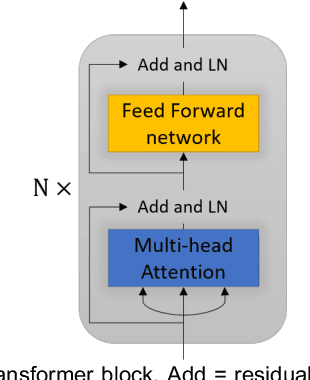
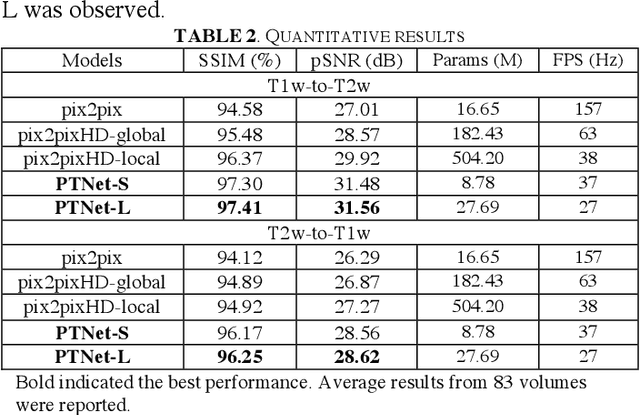
Abstract:Magnetic resonance imaging (MRI) noninvasively provides critical information about how human brain structures develop across stages of life. Developmental scientists are particularly interested in the first few years of neurodevelopment. Despite the success of MRI collection and analysis for adults, it is a challenge for researchers to collect high-quality multimodal MRIs from developing infants mainly because of their irregular sleep pattern, limited attention, inability to follow instructions to stay still, and a lack of analysis approaches. These challenges often lead to a significant reduction of usable data. To address this issue, researchers have explored various solutions to replace corrupted scans through synthesizing realistic MRIs. Among them, the convolution neural network (CNN) based generative adversarial network has demonstrated promising results and achieves state-of-the-art performance. However, adversarial training is unstable and may need careful tuning of regularization terms to stabilize the training. In this study, we introduced a novel MRI synthesis framework - Pyramid Transformer Net (PTNet). PTNet consists of transformer layers, skip-connections, and multi-scale pyramid representation. Compared with the most widely used CNN-based conditional GAN models (namely pix2pix and pix2pixHD), our model PTNet shows superior performance in terms of synthesis accuracy and model size. Notably, PTNet does not require any type of adversarial training and can be easily trained using the simple mean squared error loss.
Co-Seg: An Image Segmentation Framework Against Label Corruption
Jan 31, 2021



Abstract:Supervised deep learning performance is heavily tied to the availability of high-quality labels for training. Neural networks can gradually overfit corrupted labels if directly trained on noisy datasets, leading to severe performance degradation at test time. In this paper, we propose a novel deep learning framework, namely Co-Seg, to collaboratively train segmentation networks on datasets which include low-quality noisy labels. Our approach first trains two networks simultaneously to sift through all samples and obtain a subset with reliable labels. Then, an efficient yet easily-implemented label correction strategy is applied to enrich the reliable subset. Finally, using the updated dataset, we retrain the segmentation network to finalize its parameters. Experiments in two noisy labels scenarios demonstrate that our proposed model can achieve results comparable to those obtained from supervised learning trained on the noise-free labels. In addition, our framework can be easily implemented in any segmentation algorithm to increase its robustness to noisy labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge