Xuzhe Zhang
Contrast-Invariant Self-supervised Segmentation for Quantitative Placental MRI
May 30, 2025Abstract:Accurate placental segmentation is essential for quantitative analysis of the placenta. However, this task is particularly challenging in T2*-weighted placental imaging due to: (1) weak and inconsistent boundary contrast across individual echoes; (2) the absence of manual ground truth annotations for all echo times; and (3) motion artifacts across echoes caused by fetal and maternal movement. In this work, we propose a contrast-augmented segmentation framework that leverages complementary information across multi-echo T2*-weighted MRI to learn robust, contrast-invariant representations. Our method integrates: (i) masked autoencoding (MAE) for self-supervised pretraining on unlabeled multi-echo slices; (ii) masked pseudo-labeling (MPL) for unsupervised domain adaptation across echo times; and (iii) global-local collaboration to align fine-grained features with global anatomical context. We further introduce a semantic matching loss to encourage representation consistency across echoes of the same subject. Experiments on a clinical multi-echo placental MRI dataset demonstrate that our approach generalizes effectively across echo times and outperforms both single-echo and naive fusion baselines. To our knowledge, this is the first work to systematically exploit multi-echo T2*-weighted MRI for placental segmentation.
Robust Quantification of Percent Emphysema on CT via Domain Attention: the Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Mar 06, 2024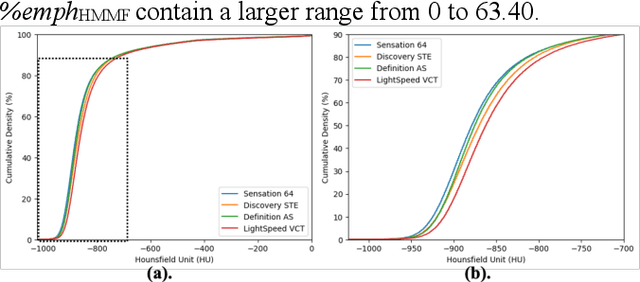
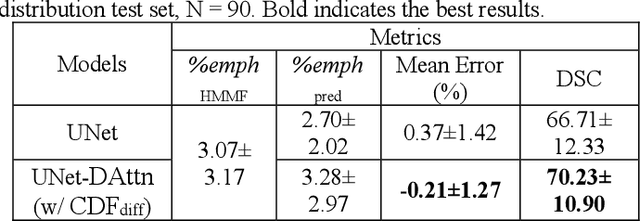
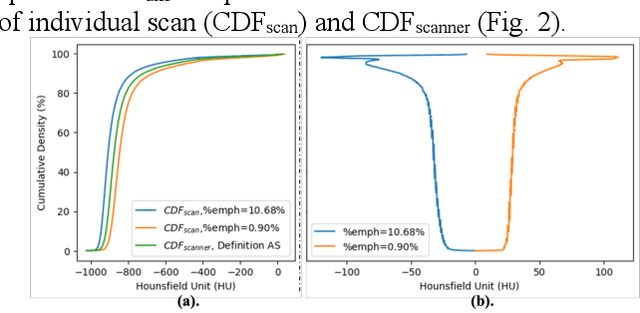
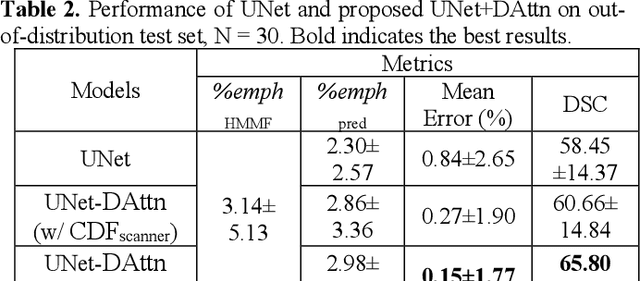
Abstract:Robust quantification of pulmonary emphysema on computed tomography (CT) remains challenging for large-scale research studies that involve scans from different scanner types and for translation to clinical scans. Existing studies have explored several directions to tackle this challenge, including density correction, noise filtering, regression, hidden Markov measure field (HMMF) model-based segmentation, and volume-adjusted lung density. Despite some promising results, previous studies either required a tedious workflow or limited opportunities for downstream emphysema subtyping, limiting efficient adaptation on a large-scale study. To alleviate this dilemma, we developed an end-to-end deep learning framework based on an existing HMMF segmentation framework. We first demonstrate that a regular UNet cannot replicate the existing HMMF results because of the lack of scanner priors. We then design a novel domain attention block to fuse image feature with quantitative scanner priors which significantly improves the results.
3D Masked Autoencoding and Pseudo-labeling for Domain Adaptive Segmentation of Heterogeneous Infant Brain MRI
Mar 16, 2023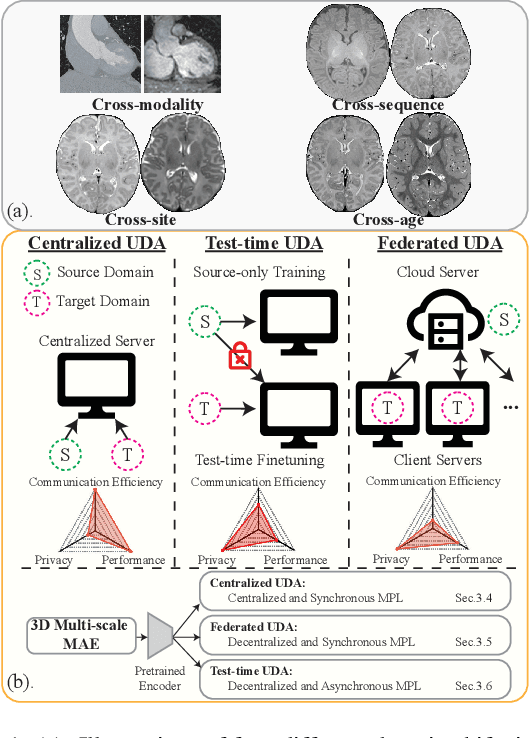
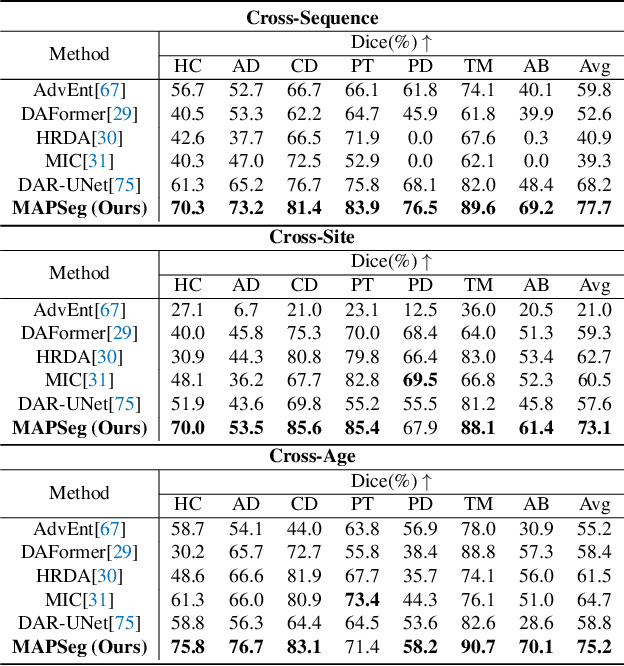
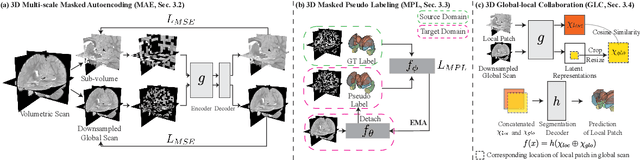

Abstract:Robust segmentation of infant brain MRI across multiple ages, modalities, and sites remains challenging due to the intrinsic heterogeneity caused by different MRI scanners, vendors, or acquisition sequences, as well as varying stages of neurodevelopment. To address this challenge, previous studies have explored domain adaptation (DA) algorithms from various perspectives, including feature alignment, entropy minimization, contrast synthesis (style transfer), and pseudo-labeling. This paper introduces a novel framework called MAPSeg (Masked Autoencoding and Pseudo-labelling Segmentation) to address the challenges of cross-age, cross-modality, and cross-site segmentation of subcortical regions in infant brain MRI. Utilizing 3D masked autoencoding as well as masked pseudo-labeling, the model is able to jointly learn from labeled source domain data and unlabeled target domain data. We evaluated our framework on expert-annotated datasets acquired from different ages and sites. MAPSeg consistently outperformed other methods, including previous state-of-the-art supervised baselines, domain generalization, and domain adaptation frameworks in segmenting subcortical regions regardless of age, modality, or acquisition site. The code and pretrained encoder will be publicly available at https://github.com/XuzheZ/MAPSeg
Recursive Refinement Network for Deformable Lung Registration between Exhale and Inhale CT Scans
Jun 14, 2021
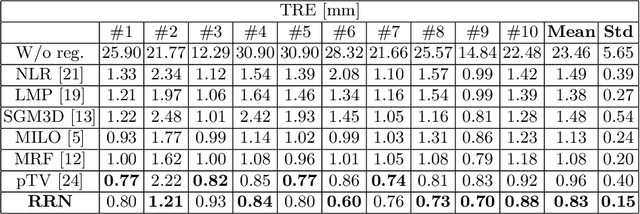
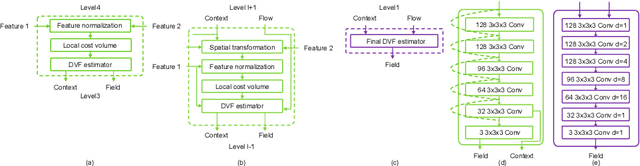
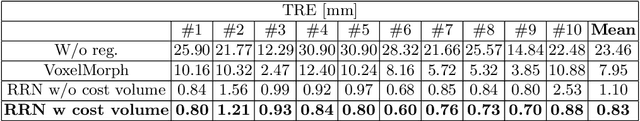
Abstract:Unsupervised learning-based medical image registration approaches have witnessed rapid development in recent years. We propose to revisit a commonly ignored while simple and well-established principle: recursive refinement of deformation vector fields across scales. We introduce a recursive refinement network (RRN) for unsupervised medical image registration, to extract multi-scale features, construct normalized local cost correlation volume and recursively refine volumetric deformation vector fields. RRN achieves state of the art performance for 3D registration of expiratory-inspiratory pairs of CT lung scans. On DirLab COPDGene dataset, RRN returns an average Target Registration Error (TRE) of 0.83 mm, which corresponds to a 13% error reduction from the best result presented in the leaderboard. In addition to comparison with conventional methods, RRN leads to 89% error reduction compared to deep-learning-based peer approaches.
PTNet: A High-Resolution Infant MRI Synthesizer Based on Transformer
May 28, 2021
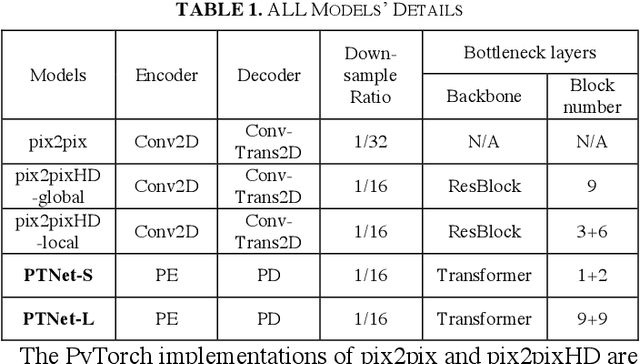
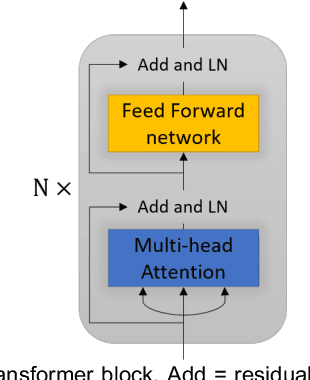
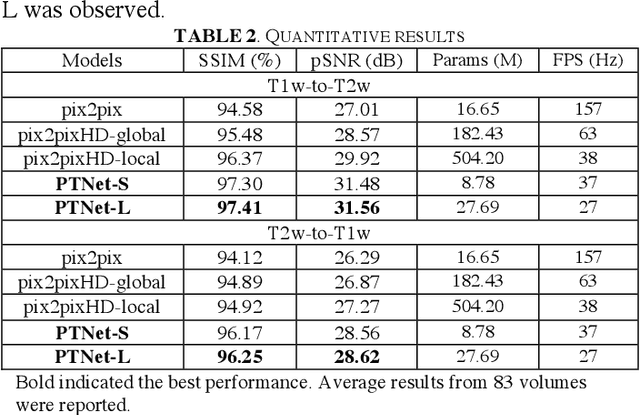
Abstract:Magnetic resonance imaging (MRI) noninvasively provides critical information about how human brain structures develop across stages of life. Developmental scientists are particularly interested in the first few years of neurodevelopment. Despite the success of MRI collection and analysis for adults, it is a challenge for researchers to collect high-quality multimodal MRIs from developing infants mainly because of their irregular sleep pattern, limited attention, inability to follow instructions to stay still, and a lack of analysis approaches. These challenges often lead to a significant reduction of usable data. To address this issue, researchers have explored various solutions to replace corrupted scans through synthesizing realistic MRIs. Among them, the convolution neural network (CNN) based generative adversarial network has demonstrated promising results and achieves state-of-the-art performance. However, adversarial training is unstable and may need careful tuning of regularization terms to stabilize the training. In this study, we introduced a novel MRI synthesis framework - Pyramid Transformer Net (PTNet). PTNet consists of transformer layers, skip-connections, and multi-scale pyramid representation. Compared with the most widely used CNN-based conditional GAN models (namely pix2pix and pix2pixHD), our model PTNet shows superior performance in terms of synthesis accuracy and model size. Notably, PTNet does not require any type of adversarial training and can be easily trained using the simple mean squared error loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge