Elsa D. Angelini
Multi-View Transformers for Airway-To-Lung Ratio Inference on Cardiac CT Scans: The C4R Study
Jan 15, 2025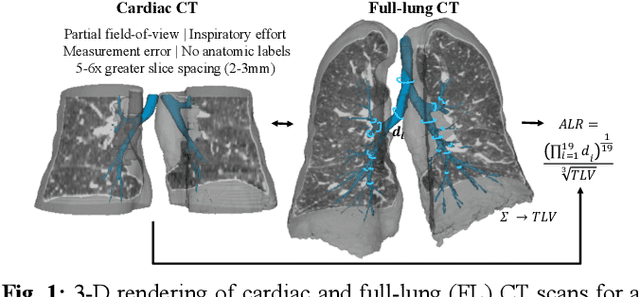
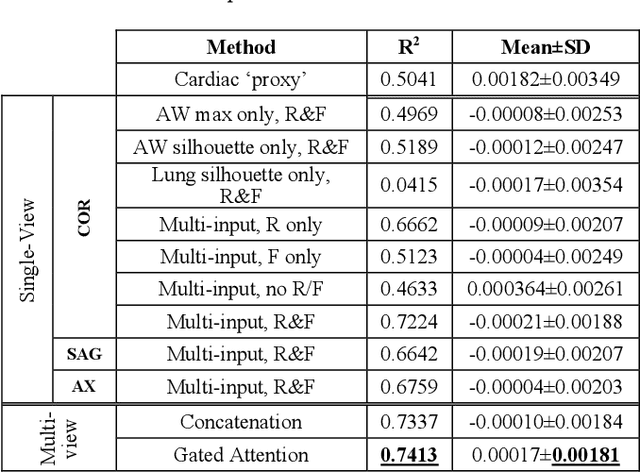
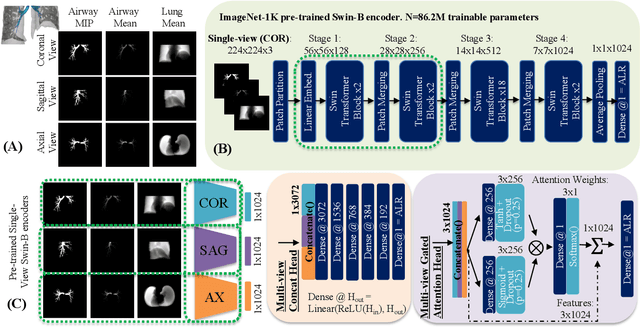
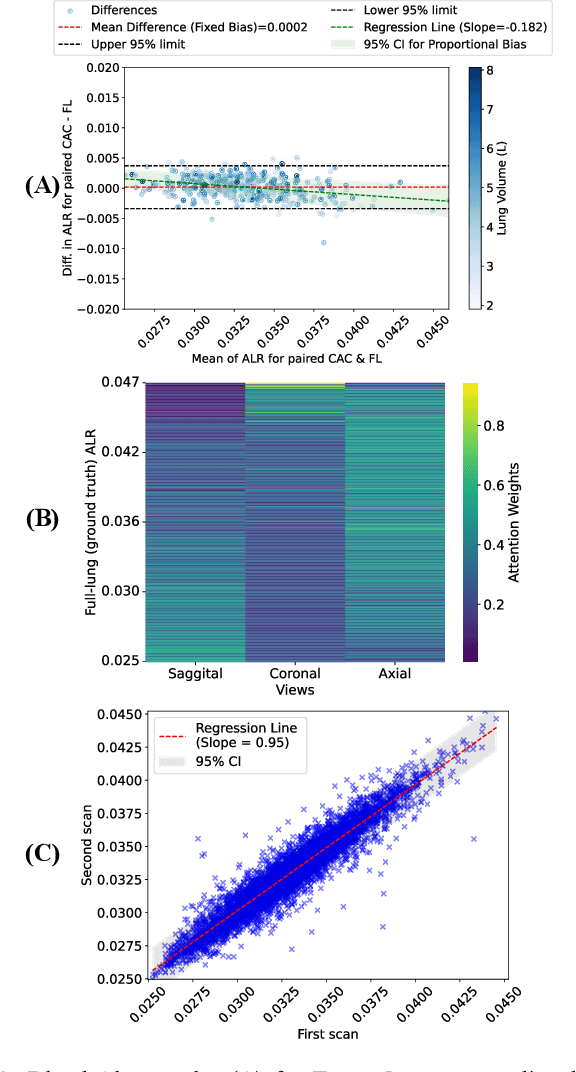
Abstract:The ratio of airway tree lumen to lung size (ALR), assessed at full inspiration on high resolution full-lung computed tomography (CT), is a major risk factor for chronic obstructive pulmonary disease (COPD). There is growing interest to infer ALR from cardiac CT images, which are widely available in epidemiological cohorts, to investigate the relationship of ALR to severe COVID-19 and post-acute sequelae of SARS-CoV-2 infection (PASC). Previously, cardiac scans included approximately 2/3 of the total lung volume with 5-6x greater slice thickness than high-resolution (HR) full-lung (FL) CT. In this study, we present a novel attention-based Multi-view Swin Transformer to infer FL ALR values from segmented cardiac CT scans. For the supervised training we exploit paired full-lung and cardiac CTs acquired in the Multi-Ethnic Study of Atherosclerosis (MESA). Our network significantly outperforms a proxy direct ALR inference on segmented cardiac CT scans and achieves accuracy and reproducibility comparable with a scan-rescan reproducibility of the FL ALR ground-truth.
Robust Quantification of Percent Emphysema on CT via Domain Attention: the Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Mar 06, 2024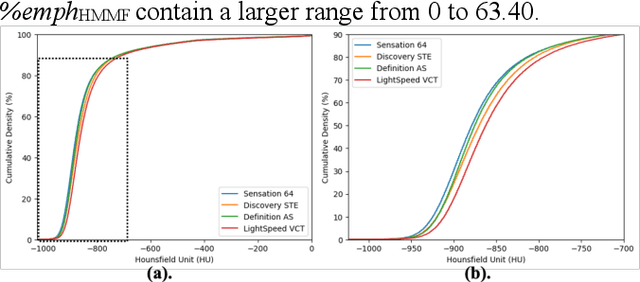
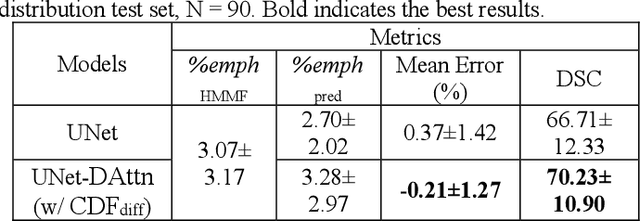
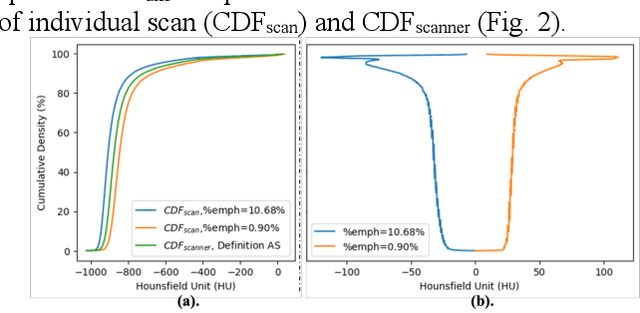
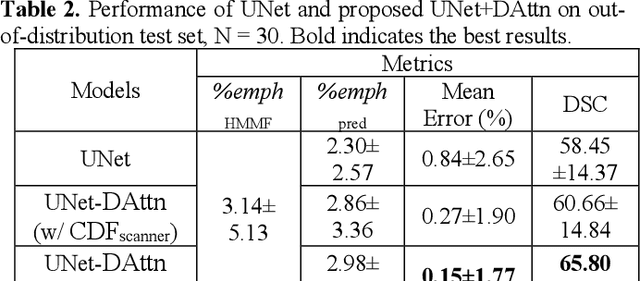
Abstract:Robust quantification of pulmonary emphysema on computed tomography (CT) remains challenging for large-scale research studies that involve scans from different scanner types and for translation to clinical scans. Existing studies have explored several directions to tackle this challenge, including density correction, noise filtering, regression, hidden Markov measure field (HMMF) model-based segmentation, and volume-adjusted lung density. Despite some promising results, previous studies either required a tedious workflow or limited opportunities for downstream emphysema subtyping, limiting efficient adaptation on a large-scale study. To alleviate this dilemma, we developed an end-to-end deep learning framework based on an existing HMMF segmentation framework. We first demonstrate that a regular UNet cannot replicate the existing HMMF results because of the lack of scanner priors. We then design a novel domain attention block to fuse image feature with quantitative scanner priors which significantly improves the results.
Robust deep labeling of radiological emphysema subtypes using squeeze and excitation convolutional neural networks: The MESA Lung and SPIROMICS Studies
Mar 01, 2024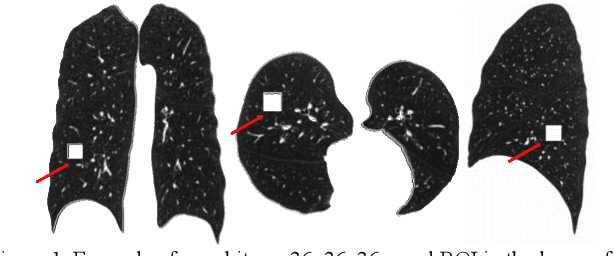
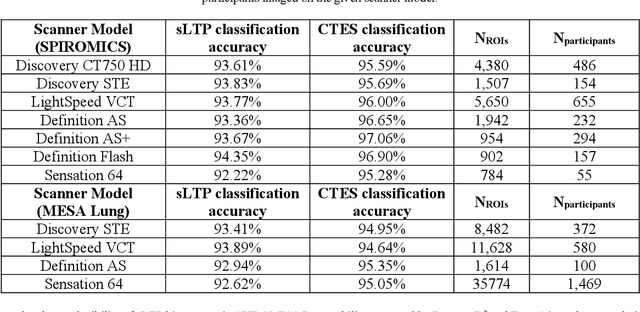
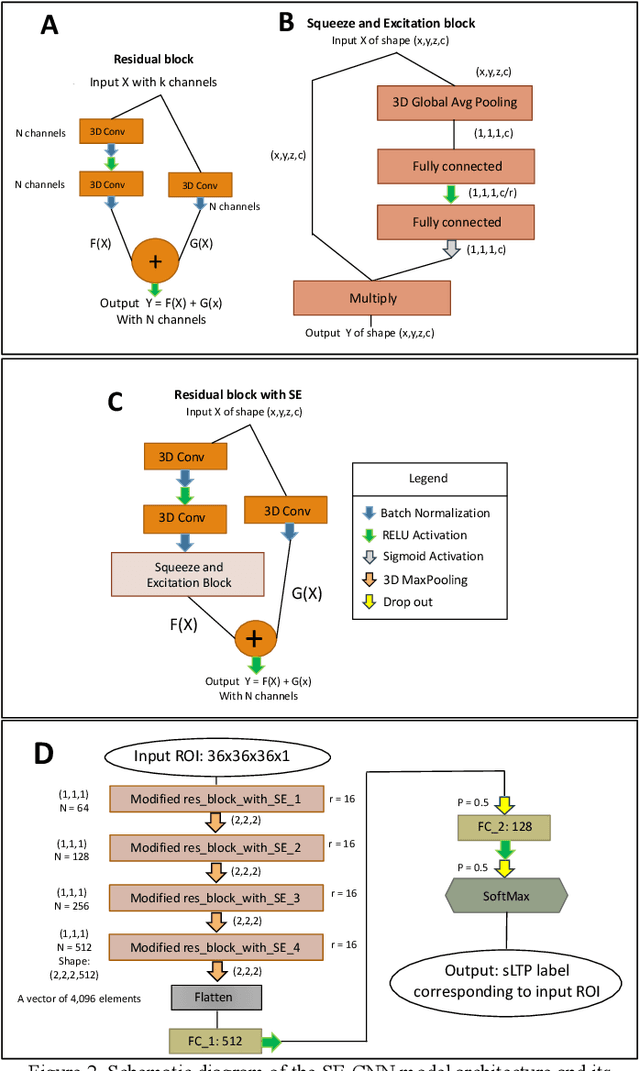
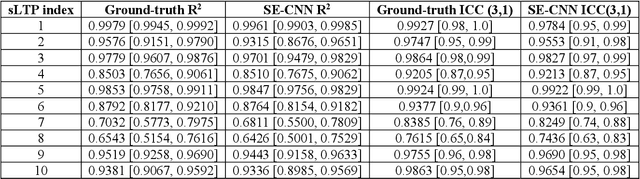
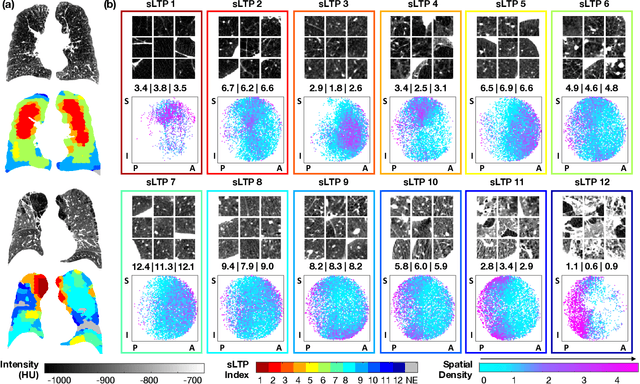
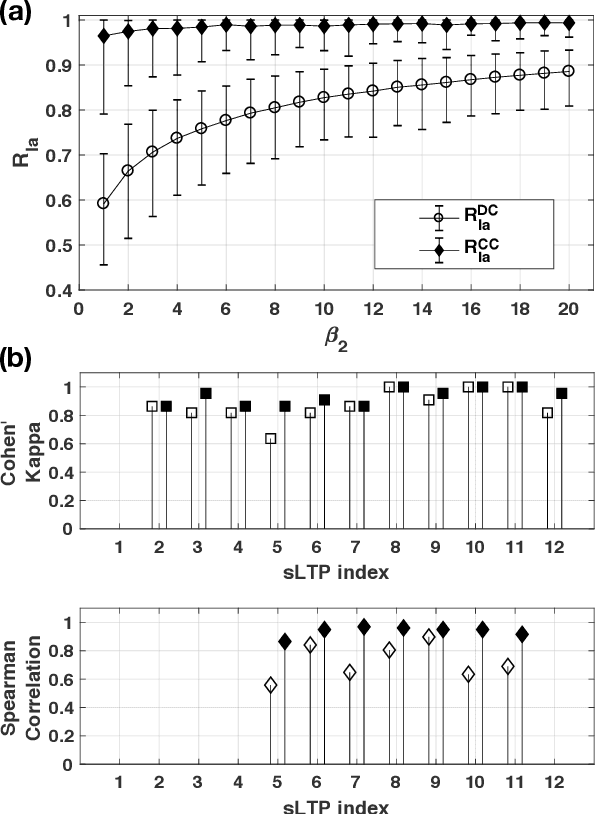
Abstract:Pulmonary emphysema, the progressive, irreversible loss of lung tissue, is conventionally categorized into three subtypes identifiable on pathology and on lung computed tomography (CT) images. Recent work has led to the unsupervised learning of ten spatially-informed lung texture patterns (sLTPs) on lung CT, representing distinct patterns of emphysematous lung parenchyma based on both textural appearance and spatial location within the lung, and which aggregate into 6 robust and reproducible CT Emphysema Subtypes (CTES). Existing methods for sLTP segmentation, however, are slow and highly sensitive to changes in CT acquisition protocol. In this work, we present a robust 3-D squeeze-and-excitation CNN for supervised classification of sLTPs and CTES on lung CT. Our results demonstrate that this model achieves accurate and reproducible sLTP segmentation on lung CTscans, across two independent cohorts and independently of scanner manufacturer and model.
Unsupervised Airway Tree Clustering with Deep Learning: The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Feb 28, 2024


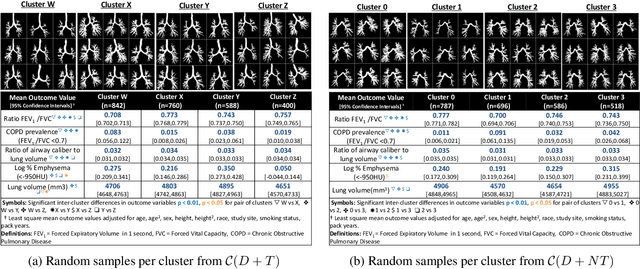
Abstract:High-resolution full lung CT scans now enable the detailed segmentation of airway trees up to the 6th branching generation. The airway binary masks display very complex tree structures that may encode biological information relevant to disease risk and yet remain challenging to exploit via traditional methods such as meshing or skeletonization. Recent clinical studies suggest that some variations in shape patterns and caliber of the human airway tree are highly associated with adverse health outcomes, including all-cause mortality and incident COPD. However, quantitative characterization of variations observed on CT segmented airway tree remain incomplete, as does our understanding of the clinical and developmental implications of such. In this work, we present an unsupervised deep-learning pipeline for feature extraction and clustering of human airway trees, learned directly from projections of 3D airway segmentations. We identify four reproducible and clinically distinct airway sub-types in the MESA Lung CT cohort.
Scribble-based fast weak-supervision and interactive corrections for segmenting whole slide images
Feb 13, 2024



Abstract:This paper proposes a dynamic interactive and weakly supervised segmentation method with minimal user interactions to address two major challenges in the segmentation of whole slide histopathology images. First, the lack of hand-annotated datasets to train algorithms. Second, the lack of interactive paradigms to enable a dialogue between the pathologist and the machine, which can be a major obstacle for use in clinical routine. We therefore propose a fast and user oriented method to bridge this gap by giving the pathologist control over the final result while limiting the number of interactions needed to achieve a good result (over 90\% on all our metrics with only 4 correction scribbles).
Novel Subtypes of Pulmonary Emphysema Based on Spatially-Informed Lung Texture Learning
Jul 09, 2020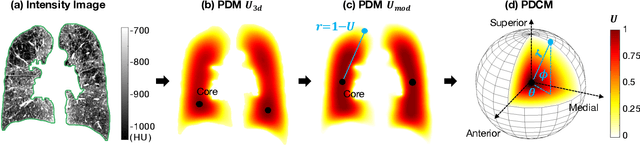
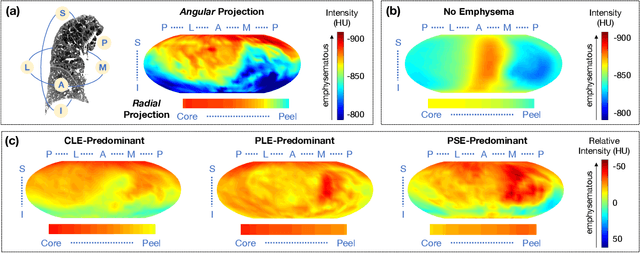
Abstract:Pulmonary emphysema overlaps considerably with chronic obstructive pulmonary disease (COPD), and is traditionally subcategorized into three subtypes previously identified on autopsy. Unsupervised learning of emphysema subtypes on computed tomography (CT) opens the way to new definitions of emphysema subtypes and eliminates the need of thorough manual labeling. However, CT-based emphysema subtypes have been limited to texture-based patterns without considering spatial location. In this work, we introduce a standardized spatial mapping of the lung for quantitative study of lung texture location, and propose a novel framework for combining spatial and texture information to discover spatially-informed lung texture patterns (sLTPs) that represent novel emphysema subtypes. Exploiting two cohorts of full-lung CT scans from the MESA COPD and EMCAP studies, we first show that our spatial mapping enables population-wide study of emphysema spatial location. We then evaluate the characteristics of the sLTPs discovered on MESA COPD, and show that they are reproducible, able to encode standard emphysema subtypes, and associated with physiological symptoms.
Discriminative analysis of the human cortex using spherical CNNs - a study on Alzheimer's disease diagnosis
Dec 19, 2018



Abstract:In neuroimaging studies, the human cortex is commonly modeled as a sphere to preserve the topological structure of the cortical surface. Cortical neuroimaging measures hence can be modeled in spherical representation. In this work, we explore analyzing the human cortex using spherical CNNs in an Alzheimer's disease (AD) classification task using cortical morphometric measures derived from structural MRI. Our results show superior performance in classifying AD versus cognitively normal and in predicting MCI progression within two years, using structural MRI information only. This work demonstrates for the first time the potential of the spherical CNNs framework in the discriminative analysis of the human cortex and could be extended to other modalities and other neurological diseases.
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules
Feb 22, 2018



Abstract:Automated detection and segmentation of pulmonary nodules on lung computed tomography (CT) scans can facilitate early lung cancer diagnosis. Existing supervised approaches for automated nodule segmentation on CT scans require voxel-based annotations for training, which are labor- and time-consuming to obtain. In this work, we propose a weakly-supervised method that generates accurate voxel-level nodule segmentation trained with image-level labels only. By adapting a convolutional neural network (CNN) trained for image classification, our proposed method learns discriminative regions from the activation maps of convolution units at different scales, and identifies the true nodule location with a novel candidate-screening framework. Experimental results on the public LIDC-IDRI dataset demonstrate that, our weakly-supervised nodule segmentation framework achieves competitive performance compared to a fully-supervised CNN-based segmentation method.
Explaining Radiological Emphysema Subtypes with Unsupervised Texture Prototypes: MESA COPD Study
Dec 05, 2016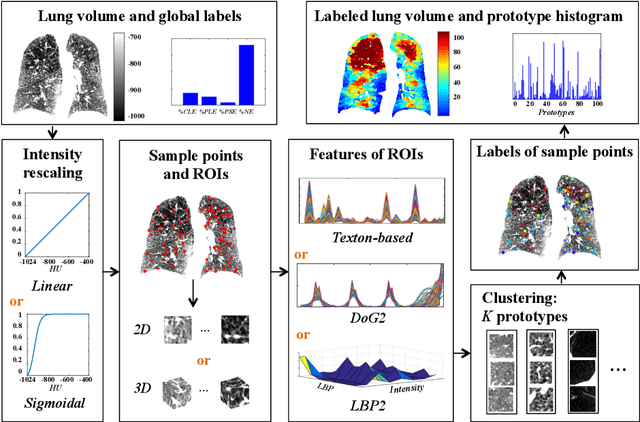
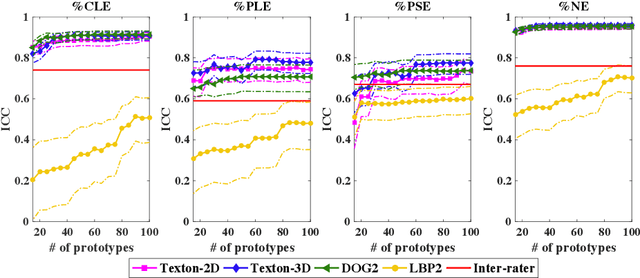
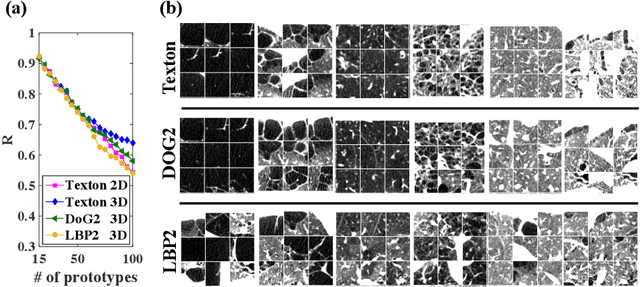

Abstract:Pulmonary emphysema is traditionally subcategorized into three subtypes, which have distinct radiological appearances on computed tomography (CT) and can help with the diagnosis of chronic obstructive pulmonary disease (COPD). Automated texture-based quantification of emphysema subtypes has been successfully implemented via supervised learning of these three emphysema subtypes. In this work, we demonstrate that unsupervised learning on a large heterogeneous database of CT scans can generate texture prototypes that are visually homogeneous and distinct, reproducible across subjects, and capable of predicting accurately the three standard radiological subtypes. These texture prototypes enable automated labeling of lung volumes, and open the way to new interpretations of lung CT scans with finer subtyping of emphysema.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge