Andrew F. Laine
Contrast-Invariant Self-supervised Segmentation for Quantitative Placental MRI
May 30, 2025Abstract:Accurate placental segmentation is essential for quantitative analysis of the placenta. However, this task is particularly challenging in T2*-weighted placental imaging due to: (1) weak and inconsistent boundary contrast across individual echoes; (2) the absence of manual ground truth annotations for all echo times; and (3) motion artifacts across echoes caused by fetal and maternal movement. In this work, we propose a contrast-augmented segmentation framework that leverages complementary information across multi-echo T2*-weighted MRI to learn robust, contrast-invariant representations. Our method integrates: (i) masked autoencoding (MAE) for self-supervised pretraining on unlabeled multi-echo slices; (ii) masked pseudo-labeling (MPL) for unsupervised domain adaptation across echo times; and (iii) global-local collaboration to align fine-grained features with global anatomical context. We further introduce a semantic matching loss to encourage representation consistency across echoes of the same subject. Experiments on a clinical multi-echo placental MRI dataset demonstrate that our approach generalizes effectively across echo times and outperforms both single-echo and naive fusion baselines. To our knowledge, this is the first work to systematically exploit multi-echo T2*-weighted MRI for placental segmentation.
Multi-View Transformers for Airway-To-Lung Ratio Inference on Cardiac CT Scans: The C4R Study
Jan 15, 2025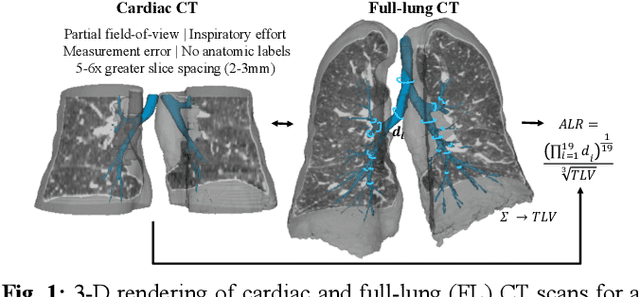
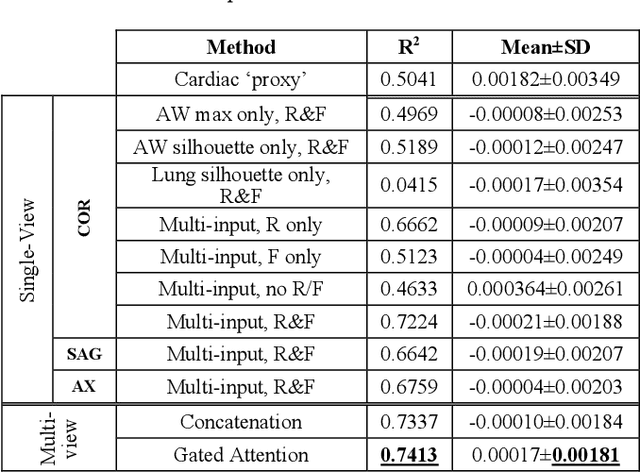
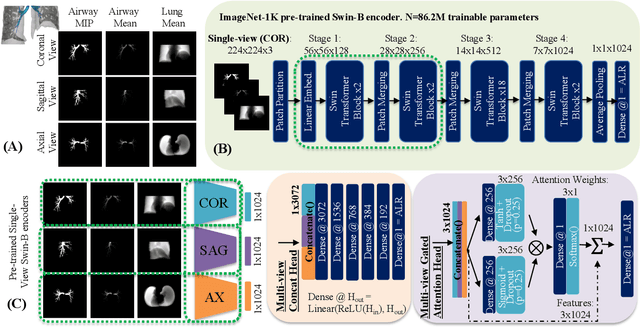
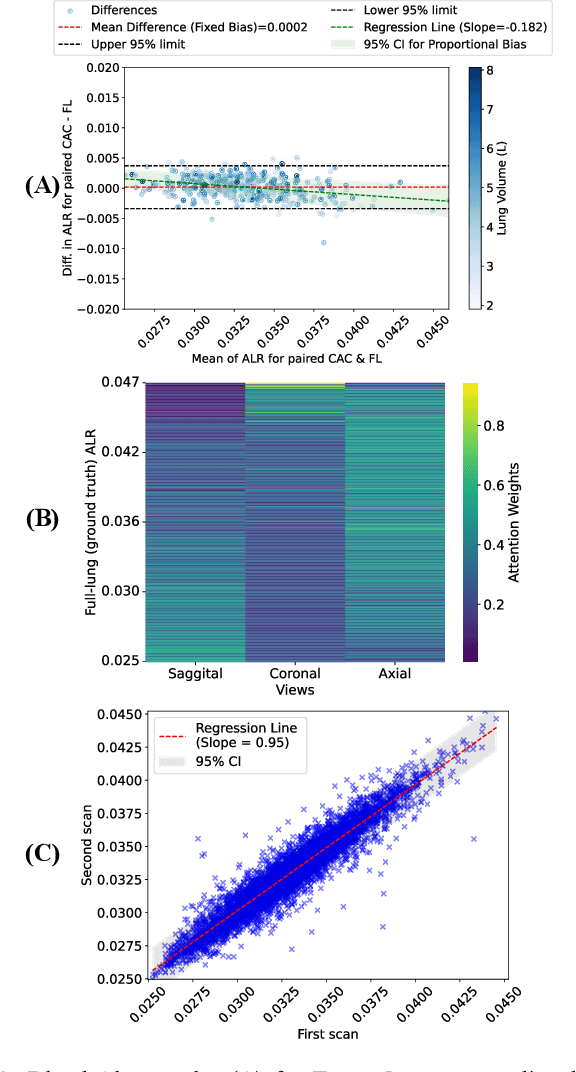
Abstract:The ratio of airway tree lumen to lung size (ALR), assessed at full inspiration on high resolution full-lung computed tomography (CT), is a major risk factor for chronic obstructive pulmonary disease (COPD). There is growing interest to infer ALR from cardiac CT images, which are widely available in epidemiological cohorts, to investigate the relationship of ALR to severe COVID-19 and post-acute sequelae of SARS-CoV-2 infection (PASC). Previously, cardiac scans included approximately 2/3 of the total lung volume with 5-6x greater slice thickness than high-resolution (HR) full-lung (FL) CT. In this study, we present a novel attention-based Multi-view Swin Transformer to infer FL ALR values from segmented cardiac CT scans. For the supervised training we exploit paired full-lung and cardiac CTs acquired in the Multi-Ethnic Study of Atherosclerosis (MESA). Our network significantly outperforms a proxy direct ALR inference on segmented cardiac CT scans and achieves accuracy and reproducibility comparable with a scan-rescan reproducibility of the FL ALR ground-truth.
Robust Quantification of Percent Emphysema on CT via Domain Attention: the Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Mar 06, 2024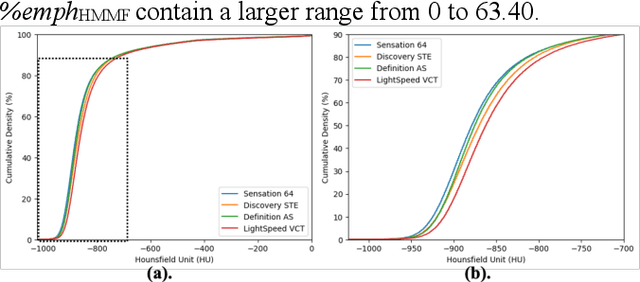
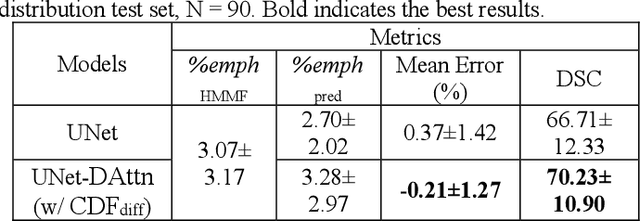
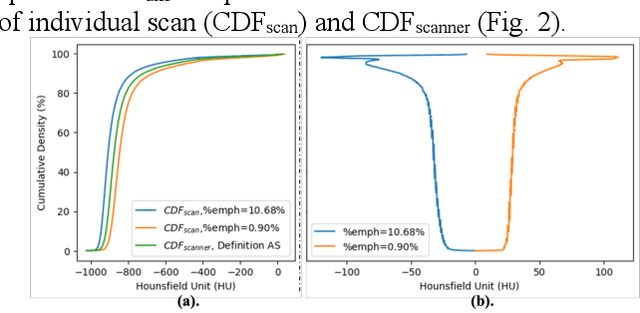
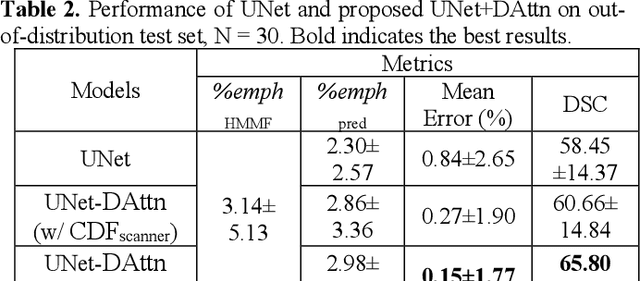
Abstract:Robust quantification of pulmonary emphysema on computed tomography (CT) remains challenging for large-scale research studies that involve scans from different scanner types and for translation to clinical scans. Existing studies have explored several directions to tackle this challenge, including density correction, noise filtering, regression, hidden Markov measure field (HMMF) model-based segmentation, and volume-adjusted lung density. Despite some promising results, previous studies either required a tedious workflow or limited opportunities for downstream emphysema subtyping, limiting efficient adaptation on a large-scale study. To alleviate this dilemma, we developed an end-to-end deep learning framework based on an existing HMMF segmentation framework. We first demonstrate that a regular UNet cannot replicate the existing HMMF results because of the lack of scanner priors. We then design a novel domain attention block to fuse image feature with quantitative scanner priors which significantly improves the results.
Robust deep labeling of radiological emphysema subtypes using squeeze and excitation convolutional neural networks: The MESA Lung and SPIROMICS Studies
Mar 01, 2024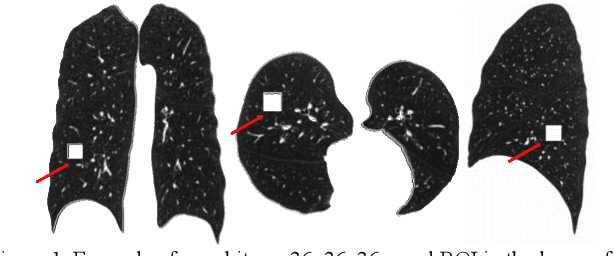
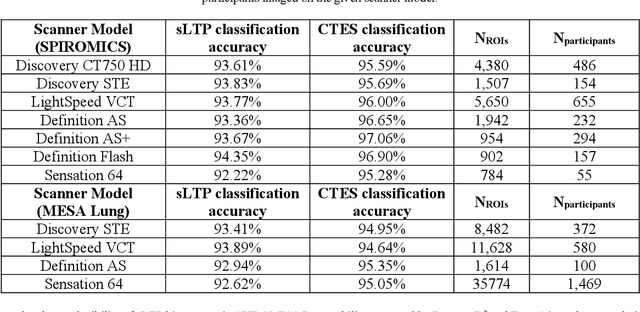
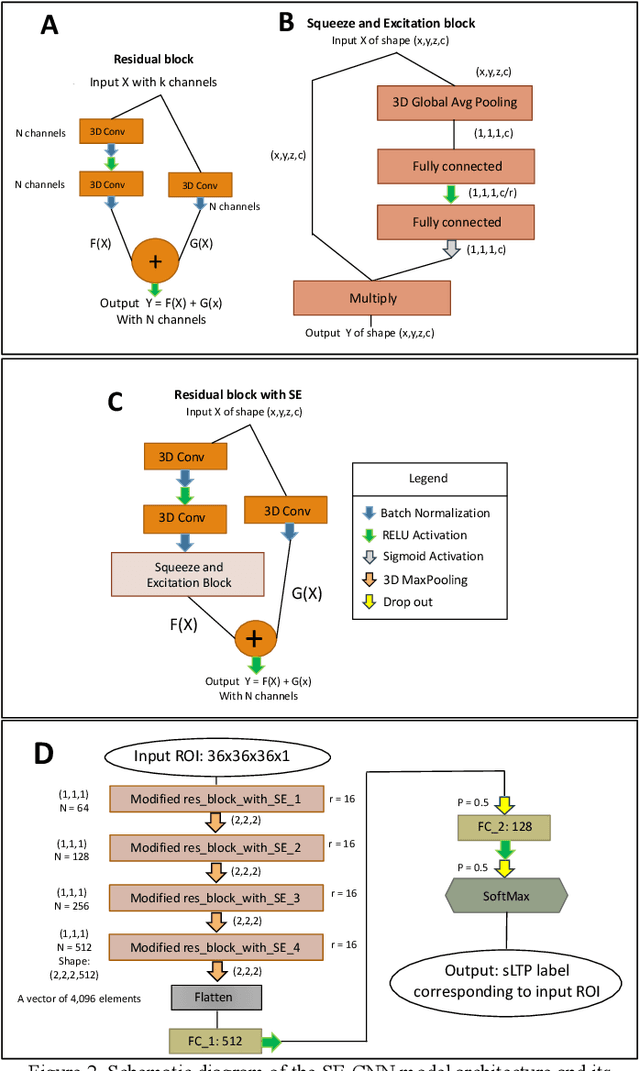
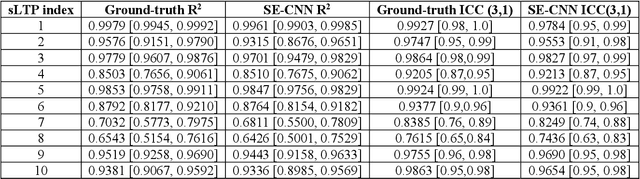
Abstract:Pulmonary emphysema, the progressive, irreversible loss of lung tissue, is conventionally categorized into three subtypes identifiable on pathology and on lung computed tomography (CT) images. Recent work has led to the unsupervised learning of ten spatially-informed lung texture patterns (sLTPs) on lung CT, representing distinct patterns of emphysematous lung parenchyma based on both textural appearance and spatial location within the lung, and which aggregate into 6 robust and reproducible CT Emphysema Subtypes (CTES). Existing methods for sLTP segmentation, however, are slow and highly sensitive to changes in CT acquisition protocol. In this work, we present a robust 3-D squeeze-and-excitation CNN for supervised classification of sLTPs and CTES on lung CT. Our results demonstrate that this model achieves accurate and reproducible sLTP segmentation on lung CTscans, across two independent cohorts and independently of scanner manufacturer and model.
Unsupervised Airway Tree Clustering with Deep Learning: The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Feb 28, 2024


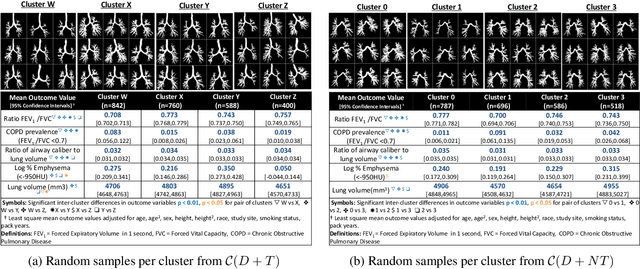
Abstract:High-resolution full lung CT scans now enable the detailed segmentation of airway trees up to the 6th branching generation. The airway binary masks display very complex tree structures that may encode biological information relevant to disease risk and yet remain challenging to exploit via traditional methods such as meshing or skeletonization. Recent clinical studies suggest that some variations in shape patterns and caliber of the human airway tree are highly associated with adverse health outcomes, including all-cause mortality and incident COPD. However, quantitative characterization of variations observed on CT segmented airway tree remain incomplete, as does our understanding of the clinical and developmental implications of such. In this work, we present an unsupervised deep-learning pipeline for feature extraction and clustering of human airway trees, learned directly from projections of 3D airway segmentations. We identify four reproducible and clinically distinct airway sub-types in the MESA Lung CT cohort.
TABSurfer: a Hybrid Deep Learning Architecture for Subcortical Segmentation
Dec 13, 2023Abstract:Subcortical segmentation remains challenging despite its important applications in quantitative structural analysis of brain MRI scans. The most accurate method, manual segmentation, is highly labor intensive, so automated tools like FreeSurfer have been adopted to handle this task. However, these traditional pipelines are slow and inefficient for processing large datasets. In this study, we propose TABSurfer, a novel 3D patch-based CNN-Transformer hybrid deep learning model designed for superior subcortical segmentation compared to existing state-of-the-art tools. To evaluate, we first demonstrate TABSurfer's consistent performance across various T1w MRI datasets with significantly shorter processing times compared to FreeSurfer. Then, we validate against manual segmentations, where TABSurfer outperforms FreeSurfer based on the manual ground truth. In each test, we also establish TABSurfer's advantage over a leading deep learning benchmark, FastSurferVINN. Together, these studies highlight TABSurfer's utility as a powerful tool for fully automated subcortical segmentation with high fidelity.
3D Masked Autoencoding and Pseudo-labeling for Domain Adaptive Segmentation of Heterogeneous Infant Brain MRI
Mar 16, 2023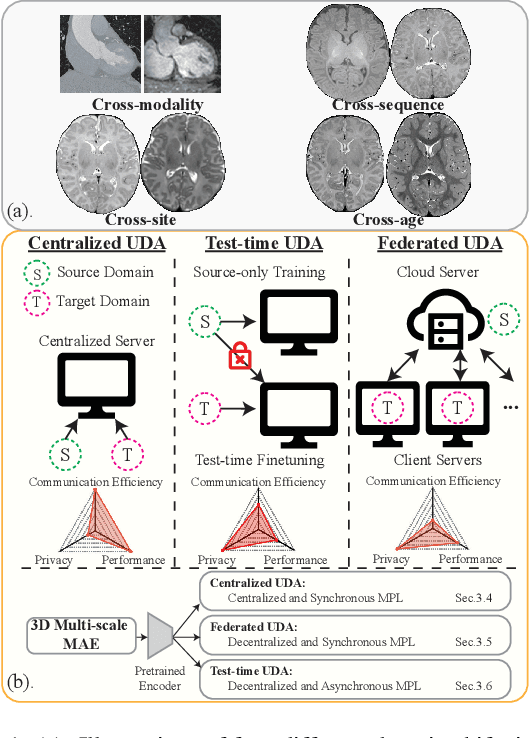
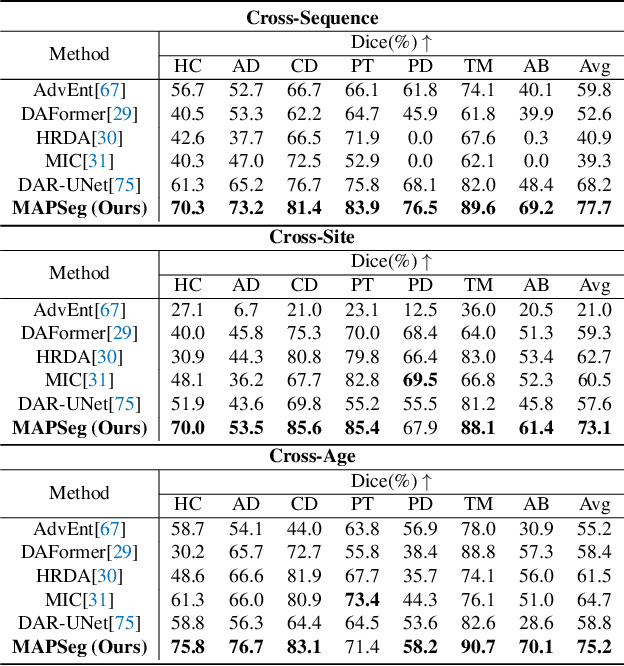
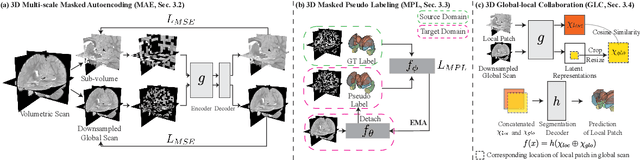

Abstract:Robust segmentation of infant brain MRI across multiple ages, modalities, and sites remains challenging due to the intrinsic heterogeneity caused by different MRI scanners, vendors, or acquisition sequences, as well as varying stages of neurodevelopment. To address this challenge, previous studies have explored domain adaptation (DA) algorithms from various perspectives, including feature alignment, entropy minimization, contrast synthesis (style transfer), and pseudo-labeling. This paper introduces a novel framework called MAPSeg (Masked Autoencoding and Pseudo-labelling Segmentation) to address the challenges of cross-age, cross-modality, and cross-site segmentation of subcortical regions in infant brain MRI. Utilizing 3D masked autoencoding as well as masked pseudo-labeling, the model is able to jointly learn from labeled source domain data and unlabeled target domain data. We evaluated our framework on expert-annotated datasets acquired from different ages and sites. MAPSeg consistently outperformed other methods, including previous state-of-the-art supervised baselines, domain generalization, and domain adaptation frameworks in segmenting subcortical regions regardless of age, modality, or acquisition site. The code and pretrained encoder will be publicly available at https://github.com/XuzheZ/MAPSeg
Improving Across-Dataset Brain Tissue Segmentation Using Transformer
Jan 21, 2022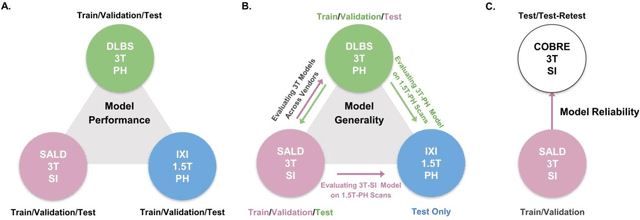
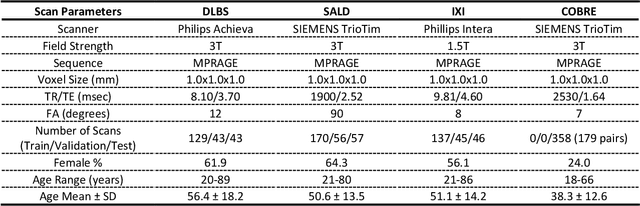
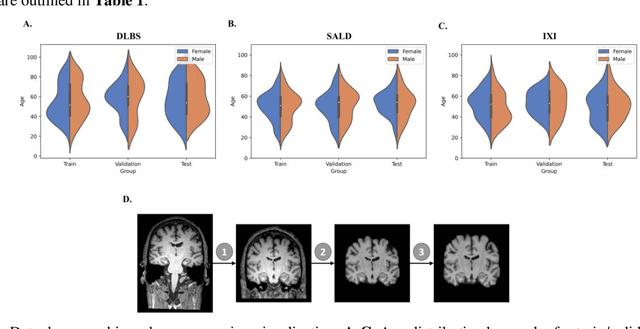
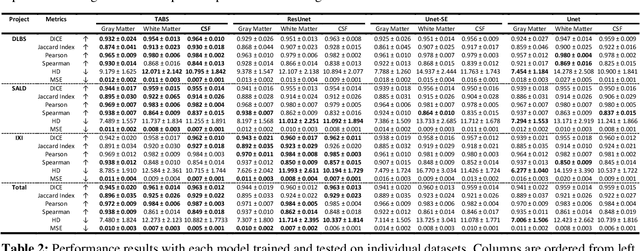
Abstract:Brain tissue segmentation has demonstrated great utility in quantifying MRI data through Voxel-Based Morphometry and highlighting subtle structural changes associated with various conditions within the brain. However, manual segmentation is highly labor-intensive, and automated approaches have struggled due to properties inherent to MRI acquisition, leaving a great need for an effective segmentation tool. Despite the recent success of deep convolutional neural networks (CNNs) for brain tissue segmentation, many such solutions do not generalize well to new datasets, which is critical for a reliable solution. Transformers have demonstrated success in natural image segmentation and have recently been applied to 3D medical image segmentation tasks due to their ability to capture long-distance relationships in the input where the local receptive fields of CNNs struggle. This study introduces a novel CNN-Transformer hybrid architecture designed for brain tissue segmentation. We validate our model's performance across four multi-site T1w MRI datasets, covering different vendors, field strengths, scan parameters, time points, and neuropsychiatric conditions. In all situations, our model achieved the greatest generality and reliability. Out method is inherently robust and can serve as a valuable tool for brain-related T1w MRI studies. The code for the TABS network is available at: https://github.com/raovish6/TABS.
Automated Multi-Process CTC Detection using Deep Learning
Sep 26, 2021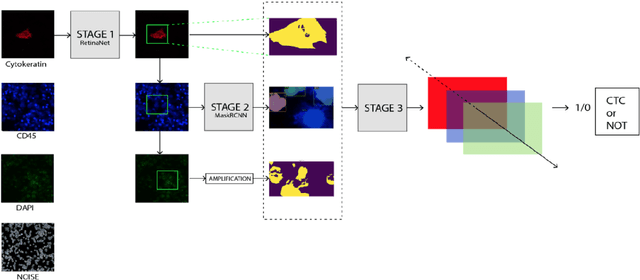
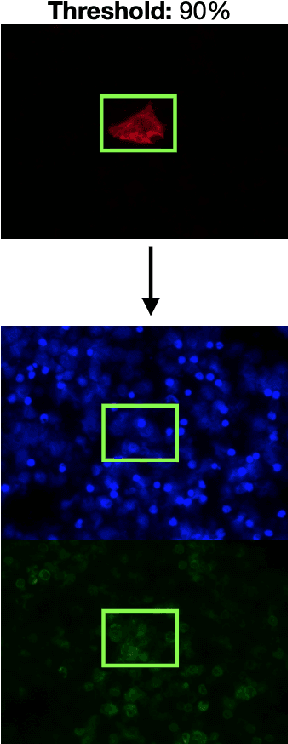
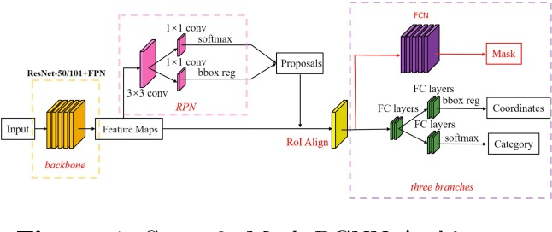
Abstract:Circulating Tumor Cells (CTCs) bear great promise as biomarkers in tumor prognosis. However, the process of identification and later enumeration of CTCs require manual labor, which is error-prone and time-consuming. The recent developments in object detection via Deep Learning using Mask-RCNNs and wider availability of pre-trained models have enabled sensitive tasks with limited data of such to be tackled with unprecedented accuracy. In this report, we present a novel 3-stage detection model for automated identification of Circulating Tumor Cells in multi-channel darkfield microscopic images comprised of: RetinaNet based identification of Cytokeratin (CK) stains, Mask-RCNN based cell detection of DAPI cell nuclei and Otsu thresholding to detect CD-45s. The training dataset is composed of 46 high variance data points, with 10 Negative and 36 Positive data points. The test set is composed of 420 negative data points. The final accuracy of the pipeline is 98.81%.
Darwin's Neural Network: AI-based Strategies for Rapid and Scalable Cell and Coronavirus Screening
Jul 22, 2020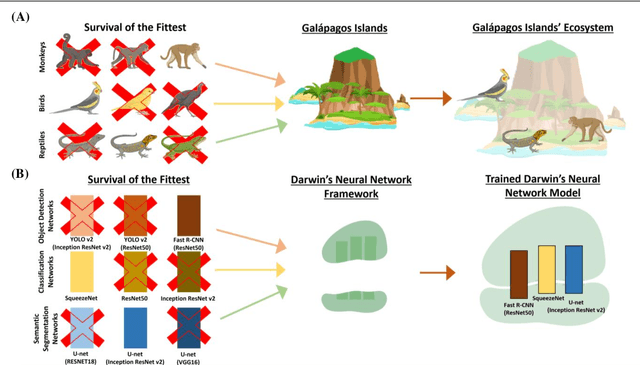
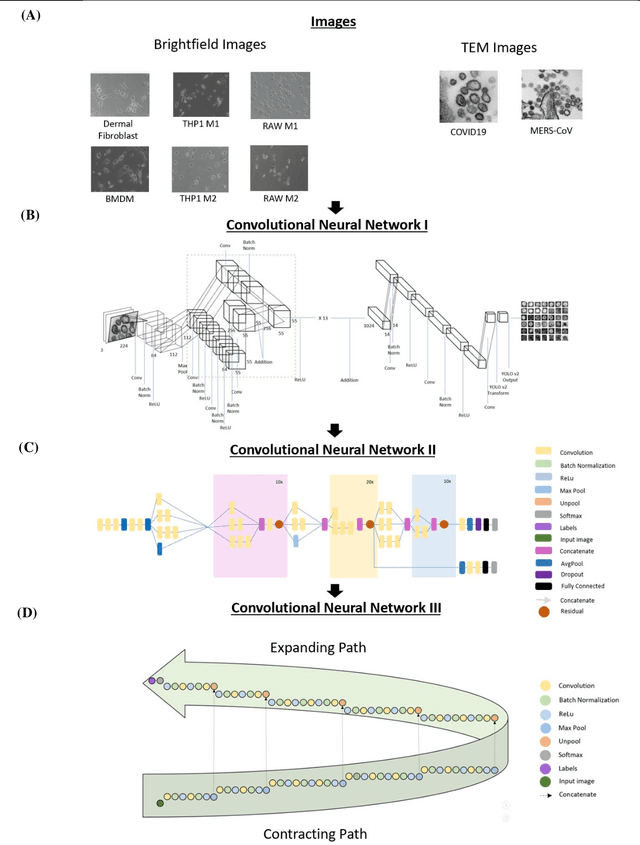
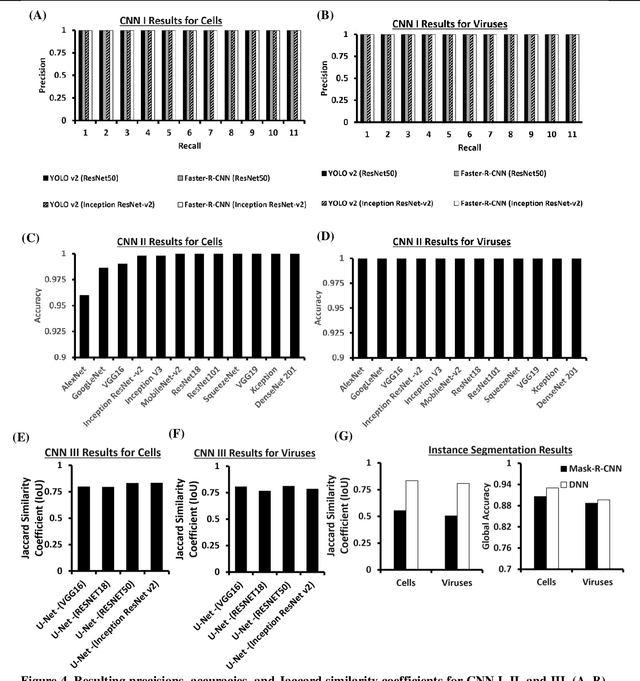
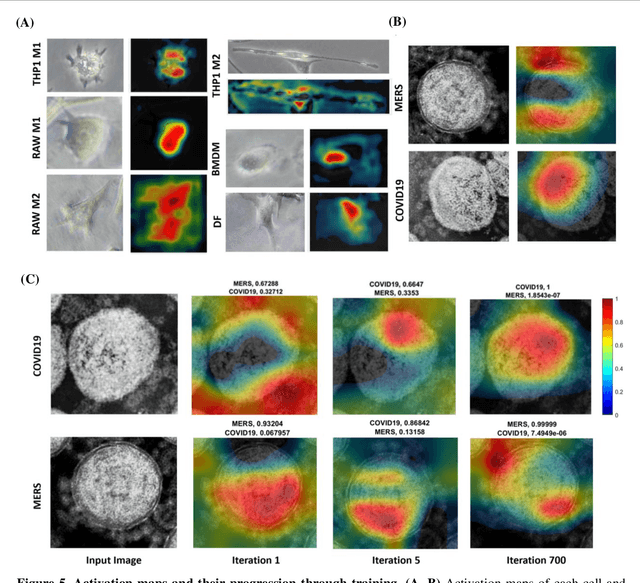
Abstract:Recent advances in the interdisciplinary scientific field of machine perception, computer vision, and biomedical engineering underpin a collection of machine learning algorithms with a remarkable ability to decipher the contents of microscope and nanoscope images. Machine learning algorithms are transforming the interpretation and analysis of microscope and nanoscope imaging data through use in conjunction with biological imaging modalities. These advances are enabling researchers to carry out real-time experiments that were previously thought to be computationally impossible. Here we adapt the theory of survival of the fittest in the field of computer vision and machine perception to introduce a new framework of multi-class instance segmentation deep learning, Darwin's Neural Network (DNN), to carry out morphometric analysis and classification of COVID19 and MERS-CoV collected in vivo and of multiple mammalian cell types in vitro.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge