Alejandro P. Comellas
Robust deep labeling of radiological emphysema subtypes using squeeze and excitation convolutional neural networks: The MESA Lung and SPIROMICS Studies
Mar 01, 2024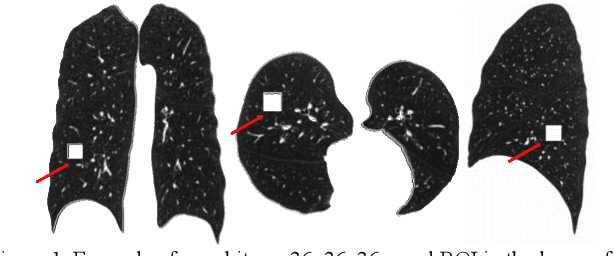
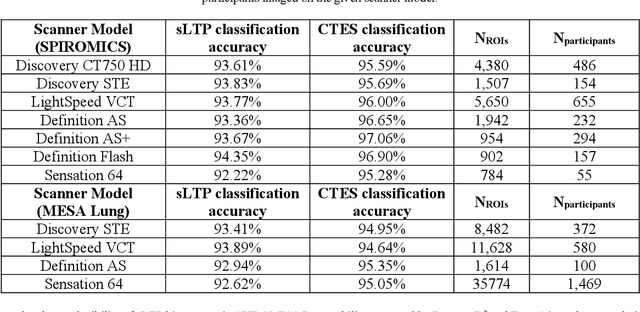
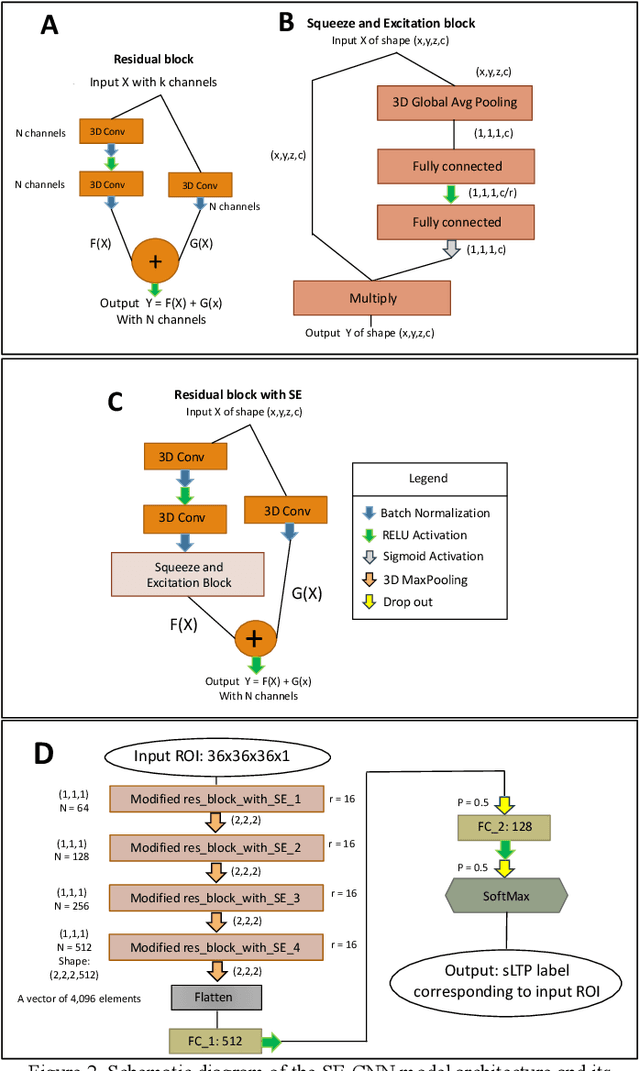
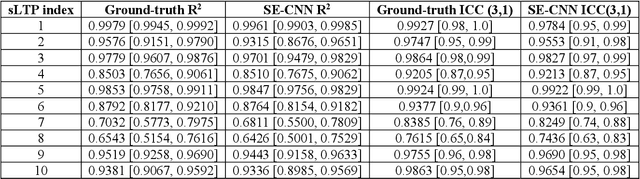
Abstract:Pulmonary emphysema, the progressive, irreversible loss of lung tissue, is conventionally categorized into three subtypes identifiable on pathology and on lung computed tomography (CT) images. Recent work has led to the unsupervised learning of ten spatially-informed lung texture patterns (sLTPs) on lung CT, representing distinct patterns of emphysematous lung parenchyma based on both textural appearance and spatial location within the lung, and which aggregate into 6 robust and reproducible CT Emphysema Subtypes (CTES). Existing methods for sLTP segmentation, however, are slow and highly sensitive to changes in CT acquisition protocol. In this work, we present a robust 3-D squeeze-and-excitation CNN for supervised classification of sLTPs and CTES on lung CT. Our results demonstrate that this model achieves accurate and reproducible sLTP segmentation on lung CTscans, across two independent cohorts and independently of scanner manufacturer and model.
MEDPSeg: End-to-end segmentation of pulmonary structures and lesions in computed tomography
Dec 04, 2023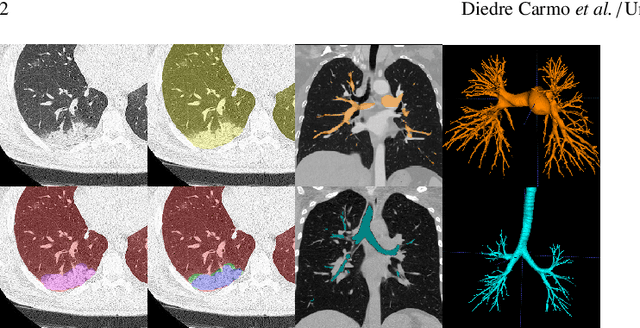
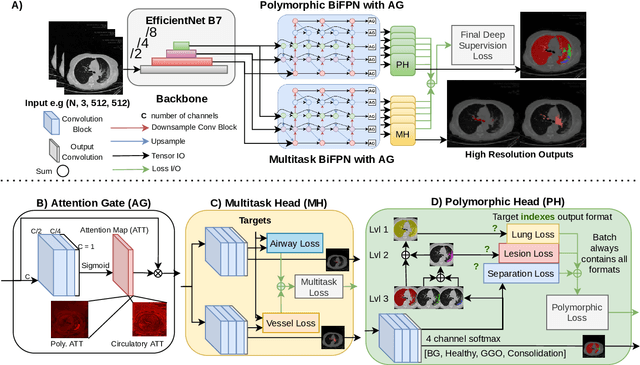

Abstract:The COVID-19 pandemic response highlighted the potential of deep learning methods in facilitating the diagnosis and prognosis of lung diseases through automated segmentation of normal and abnormal tissue in computed tomography (CT). Such methods not only have the potential to aid in clinical decision-making but also contribute to the comprehension of novel diseases. In light of the labor-intensive nature of manual segmentation for large chest CT cohorts, there is a pressing need for reliable automated approaches that enable efficient analysis of chest CT anatomy in vast research databases, especially in more scarcely annotated targets such as pneumonia consolidations. A limiting factor for the development of such methods is that most current models optimize a fixed annotation format per network output. To tackle this problem, polymorphic training is used to optimize a network with a fixed number of output channels to represent multiple hierarchical anatomic structures, indirectly optimizing more complex labels with simpler annotations. We combined over 6000 volumetric CT scans containing varying formats of manual and automated labels from different sources, and used polymorphic training along with multitask learning to develop MEDPSeg, an end-to-end method for the segmentation of lungs, airways, pulmonary artery, and lung lesions with separation of ground glass opacities, and parenchymal consolidations, all in a single forward prediction. We achieve state-of-the-art performance in multiple targets, particularly in the segmentation of ground glass opacities and consolidations, a challenging problem with limited manual annotation availability. In addition, we provide an open-source implementation with a graphical user interface at https://github.com/MICLab-Unicamp/medpseg.
Automatic segmentation of lung findings in CT and application to Long COVID
Oct 13, 2023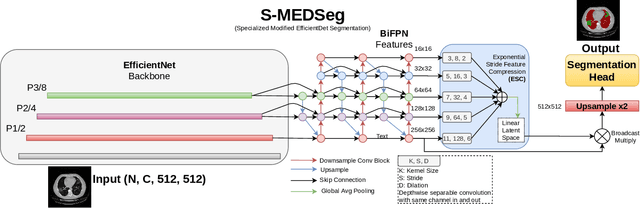
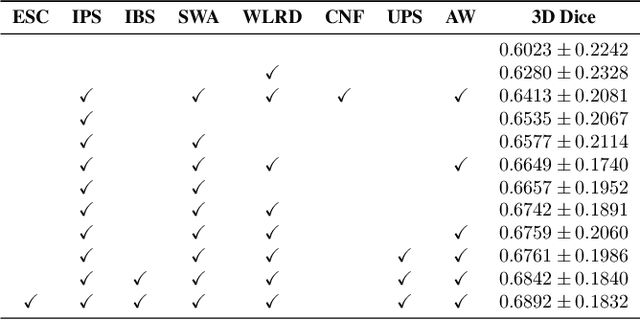
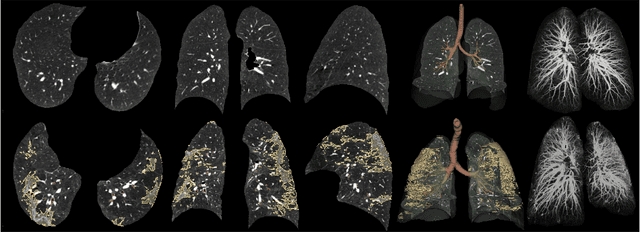
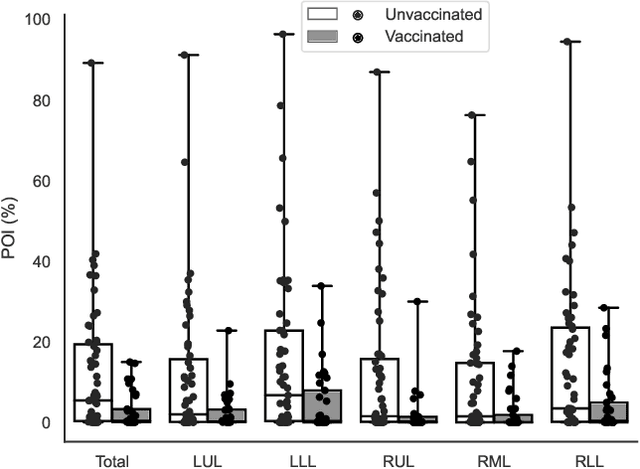
Abstract:Automated segmentation of lung abnormalities in computed tomography is an important step for diagnosing and characterizing lung disease. In this work, we improve upon a previous method and propose S-MEDSeg, a deep learning based approach for accurate segmentation of lung lesions in chest CT images. S-MEDSeg combines a pre-trained EfficientNet backbone, bidirectional feature pyramid network, and modern network advancements to achieve improved segmentation performance. A comprehensive ablation study was performed to evaluate the contribution of the proposed network modifications. The results demonstrate modifications introduced in S-MEDSeg significantly improves segmentation performance compared to the baseline approach. The proposed method is applied to an independent dataset of long COVID inpatients to study the effect of post-acute infection vaccination on extent of lung findings. Open-source code, graphical user interface and pip package are available at https://github.com/MICLab-Unicamp/medseg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge