Leticia Rittner
High-resolution segmentations of the hypothalamus and its subregions for training of segmentation models
Jun 27, 2024Abstract:Segmentation of brain structures on magnetic resonance imaging (MRI) is a highly relevant neuroimaging topic, as it is a prerequisite for different analyses such as volumetry or shape analysis. Automated segmentation facilitates the study of brain structures in larger cohorts when compared with manual segmentation, which is time-consuming. However, the development of most automated methods relies on large and manually annotated datasets, which limits the generalizability of these methods. Recently, new techniques using synthetic images have emerged, reducing the need for manual annotation. Here we provide HELM, Hypothalamic ex vivo Label Maps, a dataset composed of label maps built from publicly available ultra-high resolution ex vivo MRI from 10 whole hemispheres, which can be used to develop segmentation methods using synthetic data. The label maps are obtained with a combination of manual labels for the hypothalamic regions and automated segmentations for the rest of the brain, and mirrored to simulate entire brains. We also provide the pre-processed ex vivo scans, as this dataset can support future projects to include other structures after these are manually segmented.
H-SynEx: Using synthetic images and ultra-high resolution ex vivo MRI for hypothalamus subregion segmentation
Jan 30, 2024



Abstract:Purpose: To develop a method for automated segmentation of hypothalamus subregions informed by ultra-high resolution ex vivo magnetic resonance images (MRI), which generalizes across MRI sequences and resolutions without retraining. Materials and Methods: We trained our deep learning method, H-synEx, with synthetic images derived from label maps built from ultra-high resolution ex vivo MRI scans, which enables finer-grained manual segmentation when compared with 1mm isometric in vivo images. We validated this retrospective study using 1535 in vivo images from six datasets and six MRI sequences. The quantitative evaluation used the Dice Coefficient (DC) and Average Hausdorff distance (AVD). Statistical analysis compared hypothalamic subregion volumes in controls, Alzheimer's disease (AD), and behavioral variant frontotemporal dementia (bvFTD) subjects using the area under the curve (AUC) and Wilcoxon rank sum test. Results: H-SynEx can segment the hypothalamus across various MRI sequences, encompassing FLAIR sequences with significant slice spacing (5mm). Using hypothalamic volumes on T1w images to distinguish control from AD and bvFTD patients, we observed AUC values of 0.74 and 0.79 respectively. Additionally, AUC=0.66 was found for volume variation on FLAIR scans when comparing control and non-patients. Conclusion: Our results show that H-SynEx successfully leverages information from ultra-high resolution scans to segment in vivo from different MRI sequences such as T1w, T2w, PD, qT1, FA, and FLAIR. We also found that our automated segmentation was able to discriminate controls versus patients on FLAIR images with 5mm spacing. H-SynEx is openly available at https://github.com/liviamarodrigues/hsynex.
Spectro-ViT: A Vision Transformer Model for GABA-edited MRS Reconstruction Using Spectrograms
Nov 26, 2023Abstract:Purpose: To investigate the use of a Vision Transformer (ViT) to reconstruct/denoise GABA-edited magnetic resonance spectroscopy (MRS) from a quarter of the typically acquired number of transients using spectrograms. Theory and Methods: A quarter of the typically acquired number of transients collected in GABA-edited MRS scans are pre-processed and converted to a spectrogram image representation using the Short-Time Fourier Transform (STFT). The image representation of the data allows the adaptation of a pre-trained ViT for reconstructing GABA-edited MRS spectra (Spectro-ViT). The Spectro-ViT is fine-tuned and then tested using \textit{in vivo} GABA-edited MRS data. The Spectro-ViT performance is compared against other models in the literature using spectral quality metrics and estimated metabolite concentration values. Results: The Spectro-ViT model significantly outperformed all other models in four out of five quantitative metrics (mean squared error, shape score, GABA+/water fit error, and full width at half maximum). The metabolite concentrations estimated (GABA+/water, GABA+/Cr, and Glx/water) were consistent with the metabolite concentrations estimated using typical GABA-edited MRS scans reconstructed with the full amount of typically collected transients. Conclusion: The proposed Spectro-ViT model achieved state-of-the-art results in reconstructing GABA-edited MRS, and the results indicate these scans could be up to four times faster.
Automatic segmentation of lung findings in CT and application to Long COVID
Oct 13, 2023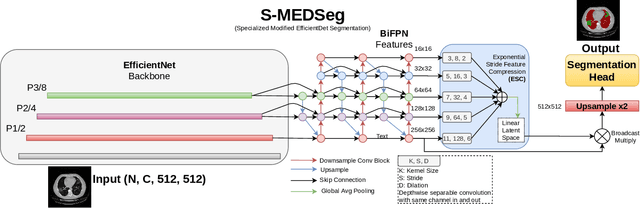
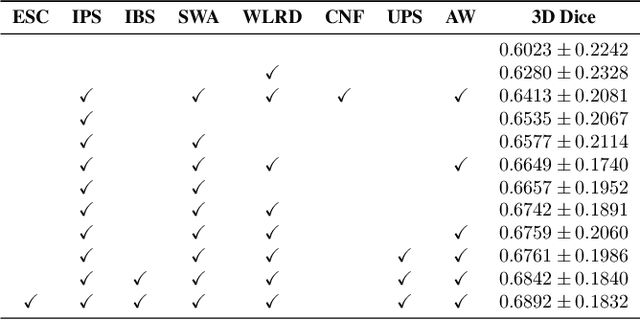
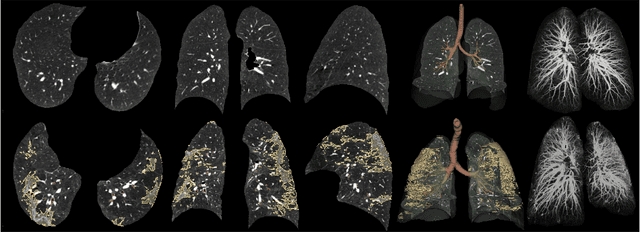
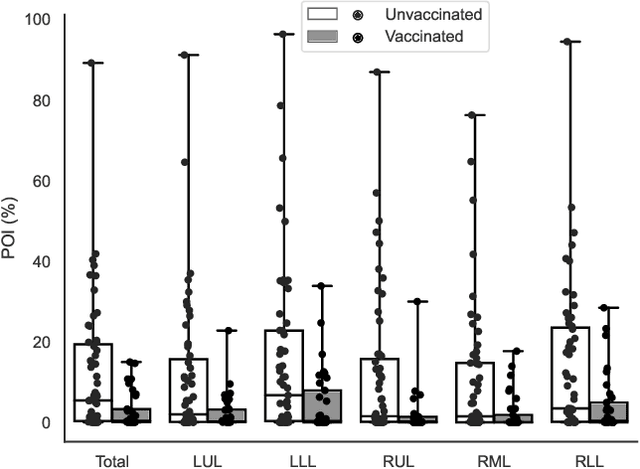
Abstract:Automated segmentation of lung abnormalities in computed tomography is an important step for diagnosing and characterizing lung disease. In this work, we improve upon a previous method and propose S-MEDSeg, a deep learning based approach for accurate segmentation of lung lesions in chest CT images. S-MEDSeg combines a pre-trained EfficientNet backbone, bidirectional feature pyramid network, and modern network advancements to achieve improved segmentation performance. A comprehensive ablation study was performed to evaluate the contribution of the proposed network modifications. The results demonstrate modifications introduced in S-MEDSeg significantly improves segmentation performance compared to the baseline approach. The proposed method is applied to an independent dataset of long COVID inpatients to study the effect of post-acute infection vaccination on extent of lung findings. Open-source code, graphical user interface and pip package are available at https://github.com/MICLab-Unicamp/medseg.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Open-source tool for Airway Segmentation in Computed Tomography using 2.5D Modified EfficientDet: Contribution to the ATM22 Challenge
Oct 03, 2022

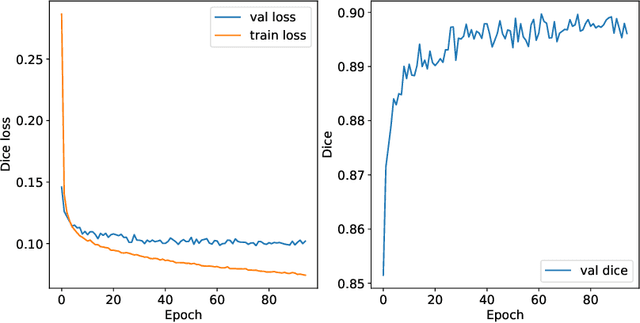

Abstract:Airway segmentation in computed tomography images can be used to analyze pulmonary diseases, however, manual segmentation is labor intensive and relies on expert knowledge. This manuscript details our contribution to MICCAI's 2022 Airway Tree Modelling challenge, a competition of fully automated methods for airway segmentation. We employed a previously developed deep learning architecture based on a modified EfficientDet (MEDSeg), training from scratch for binary airway segmentation using the provided annotations. Our method achieved 90.72 Dice in internal validation, 95.52 Dice on external validation, and 93.49 Dice in the final test phase, while not being specifically designed or tuned for airway segmentation. Open source code and a pip package for predictions with our model and trained weights are in https://github.com/MICLab-Unicamp/medseg.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019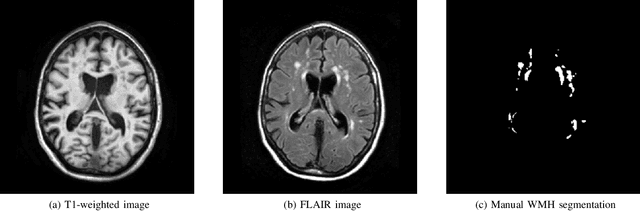
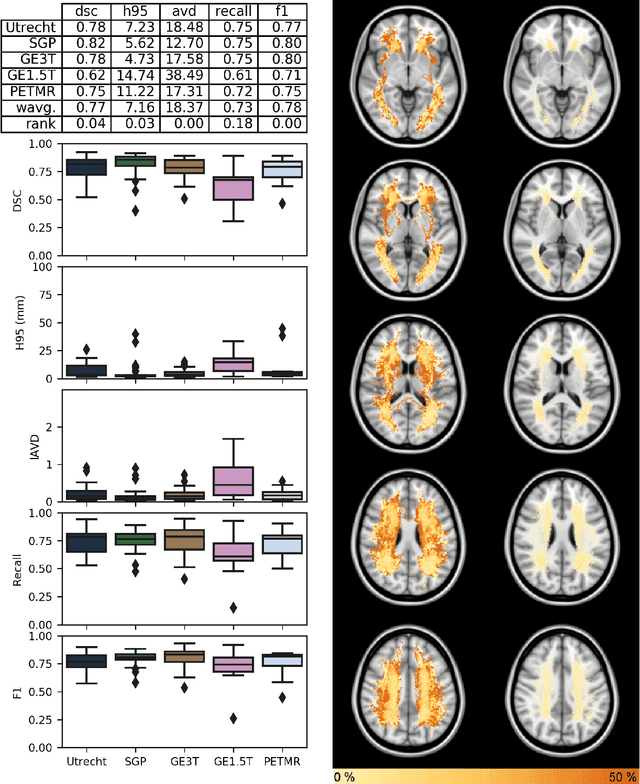
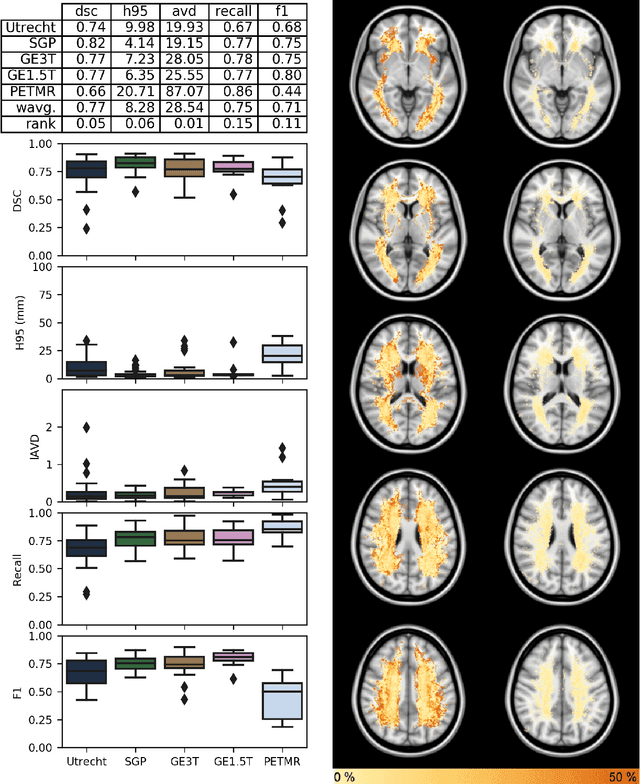
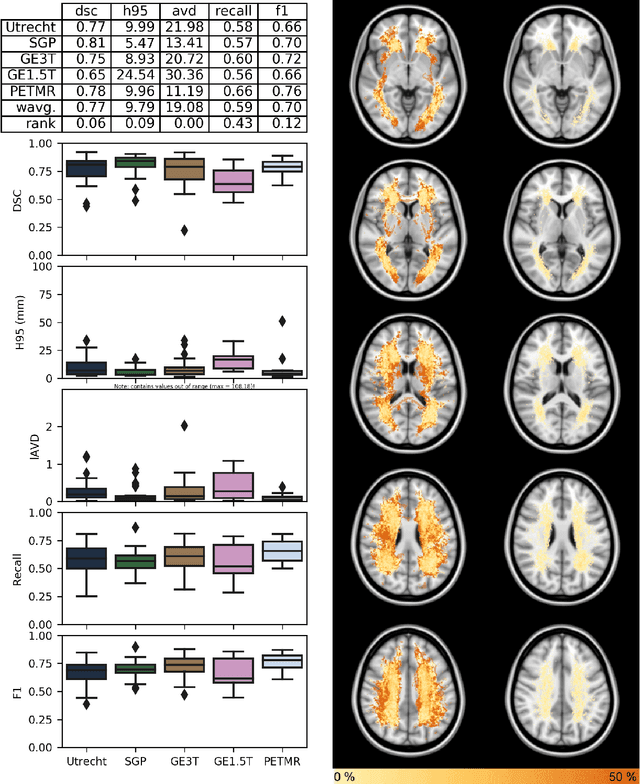
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Convolutional Neural Networks for Skull-stripping in Brain MR Imaging using Consensus-based Silver standard Masks
Apr 13, 2018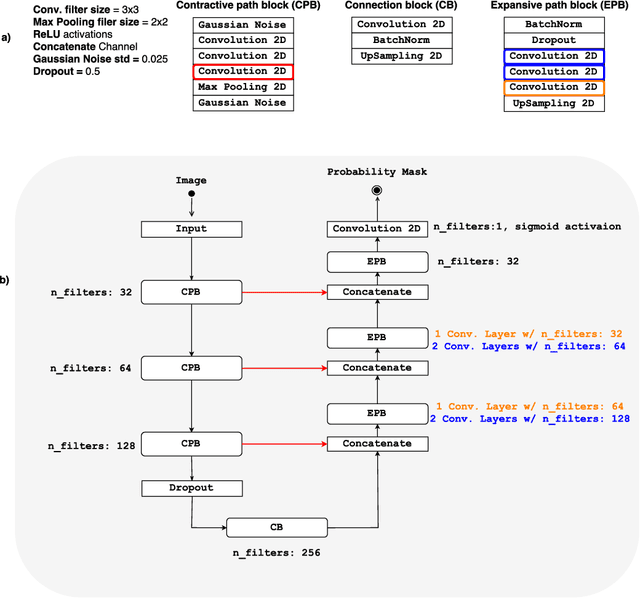
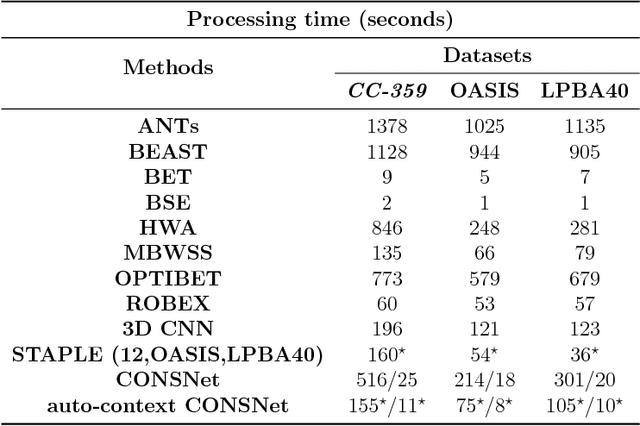
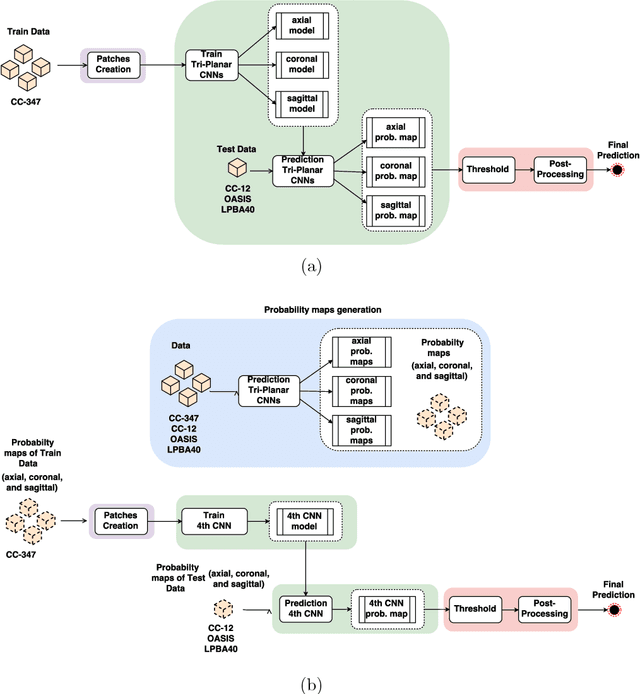
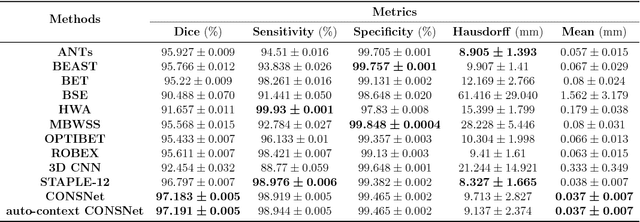
Abstract:Convolutional neural networks (CNN) for medical imaging are constrained by the number of annotated data required in the training stage. Usually, manual annotation is considered to be the "gold standard". However, medical imaging datasets that include expert manual segmentation are scarce as this step is time-consuming, and therefore expensive. Moreover, single-rater manual annotation is most often used in data-driven approaches making the network optimal with respect to only that single expert. In this work, we propose a CNN for brain extraction in magnetic resonance (MR) imaging, that is fully trained with what we refer to as silver standard masks. Our method consists of 1) developing a dataset with "silver standard" masks as input, and implementing both 2) a tri-planar method using parallel 2D U-Net-based CNNs (referred to as CONSNet) and 3) an auto-context implementation of CONSNet. The term CONSNet refers to our integrated approach, i.e., training with silver standard masks and using a 2D U-Net-based architecture. Our results showed that we outperformed (i.e., larger Dice coefficients) the current state-of-the-art SS methods. Our use of silver standard masks reduced the cost of manual annotation, decreased inter-intra-rater variability, and avoided CNN segmentation super-specialization towards one specific manual annotation guideline that can occur when gold standard masks are used. Moreover, the usage of silver standard masks greatly enlarges the volume of input annotated data because we can relatively easily generate labels for unlabeled data. In addition, our method has the advantage that, once trained, it takes only a few seconds to process a typical brain image volume using modern hardware, such as a high-end graphics processing unit. In contrast, many of the other competitive methods have processing times in the order of minutes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge