Joseph M. Reinhardt
Uncertainty-Aware Test-Time Adaptation for Inverse Consistent Diffeomorphic Lung Image Registration
Nov 12, 2024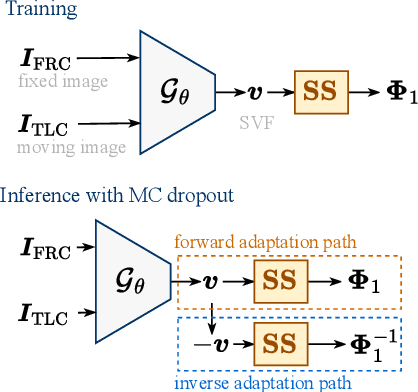
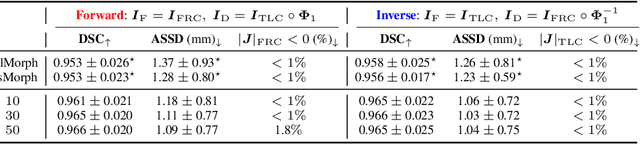
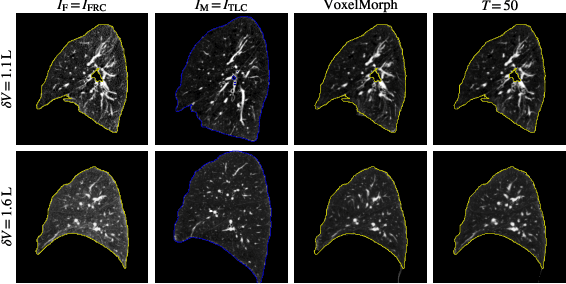
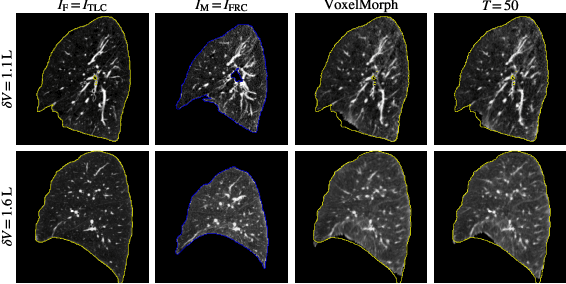
Abstract:Diffeomorphic deformable image registration ensures smooth invertible transformations across inspiratory and expiratory chest CT scans. Yet, in practice, deep learning-based diffeomorphic methods struggle to capture large deformations between inspiratory and expiratory volumes, and therefore lack inverse consistency. Existing methods also fail to account for model uncertainty, which can be useful for improving performance. We propose an uncertainty-aware test-time adaptation framework for inverse consistent diffeomorphic lung registration. Our method uses Monte Carlo (MC) dropout to estimate spatial uncertainty that is used to improve model performance. We train and evaluate our method for inspiratory-to-expiratory CT registration on a large cohort of 675 subjects from the COPDGene study, achieving a higher Dice similarity coefficient (DSC) between the lung boundaries (0.966) compared to both VoxelMorph (0.953) and TransMorph (0.953). Our method demonstrates consistent improvements in the inverse registration direction as well with an overall DSC of 0.966, higher than VoxelMorph (0.958) and TransMorph (0.956). Paired t-tests indicate statistically significant improvements.
MEDPSeg: End-to-end segmentation of pulmonary structures and lesions in computed tomography
Dec 04, 2023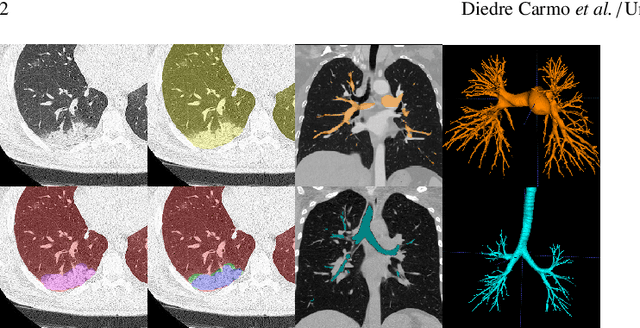
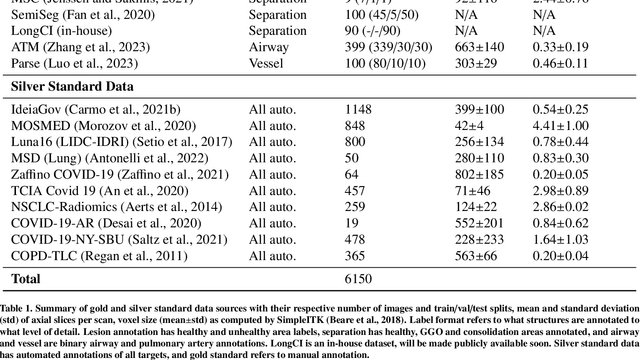
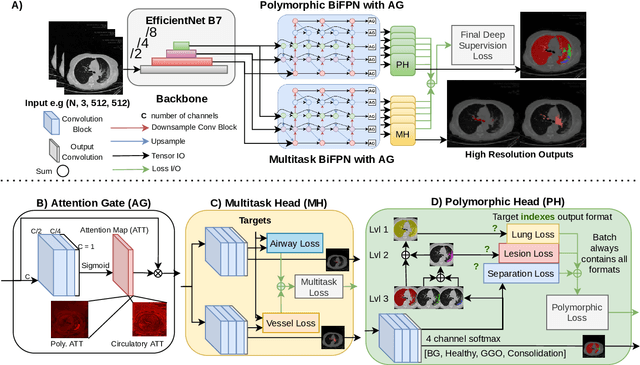

Abstract:The COVID-19 pandemic response highlighted the potential of deep learning methods in facilitating the diagnosis and prognosis of lung diseases through automated segmentation of normal and abnormal tissue in computed tomography (CT). Such methods not only have the potential to aid in clinical decision-making but also contribute to the comprehension of novel diseases. In light of the labor-intensive nature of manual segmentation for large chest CT cohorts, there is a pressing need for reliable automated approaches that enable efficient analysis of chest CT anatomy in vast research databases, especially in more scarcely annotated targets such as pneumonia consolidations. A limiting factor for the development of such methods is that most current models optimize a fixed annotation format per network output. To tackle this problem, polymorphic training is used to optimize a network with a fixed number of output channels to represent multiple hierarchical anatomic structures, indirectly optimizing more complex labels with simpler annotations. We combined over 6000 volumetric CT scans containing varying formats of manual and automated labels from different sources, and used polymorphic training along with multitask learning to develop MEDPSeg, an end-to-end method for the segmentation of lungs, airways, pulmonary artery, and lung lesions with separation of ground glass opacities, and parenchymal consolidations, all in a single forward prediction. We achieve state-of-the-art performance in multiple targets, particularly in the segmentation of ground glass opacities and consolidations, a challenging problem with limited manual annotation availability. In addition, we provide an open-source implementation with a graphical user interface at https://github.com/MICLab-Unicamp/medpseg.
Automatic segmentation of lung findings in CT and application to Long COVID
Oct 13, 2023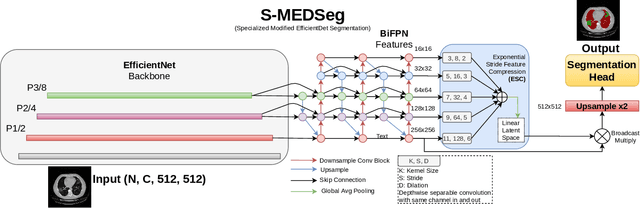
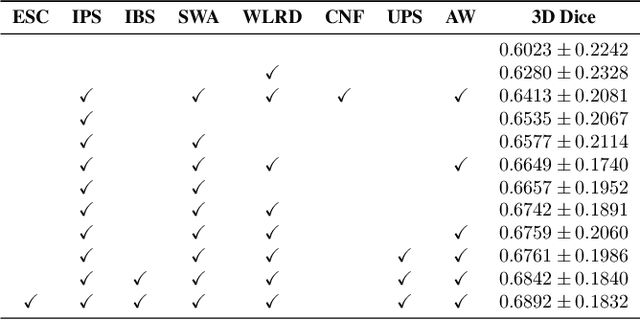
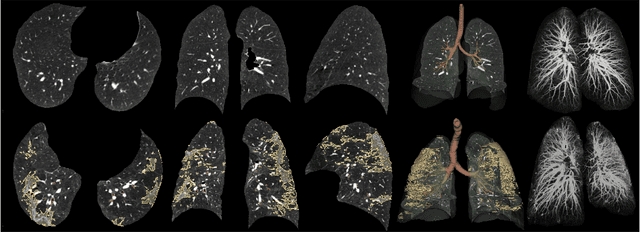
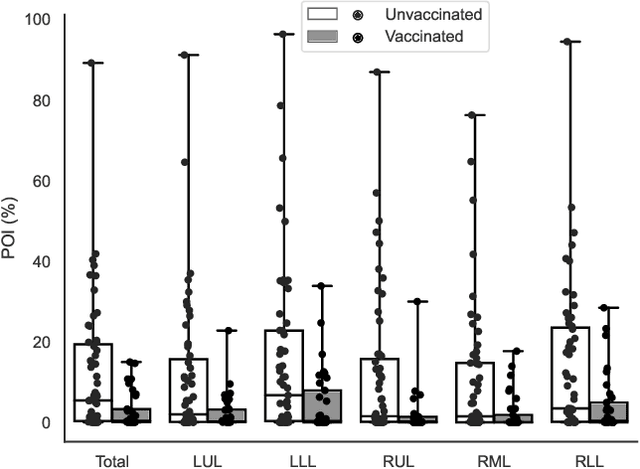
Abstract:Automated segmentation of lung abnormalities in computed tomography is an important step for diagnosing and characterizing lung disease. In this work, we improve upon a previous method and propose S-MEDSeg, a deep learning based approach for accurate segmentation of lung lesions in chest CT images. S-MEDSeg combines a pre-trained EfficientNet backbone, bidirectional feature pyramid network, and modern network advancements to achieve improved segmentation performance. A comprehensive ablation study was performed to evaluate the contribution of the proposed network modifications. The results demonstrate modifications introduced in S-MEDSeg significantly improves segmentation performance compared to the baseline approach. The proposed method is applied to an independent dataset of long COVID inpatients to study the effect of post-acute infection vaccination on extent of lung findings. Open-source code, graphical user interface and pip package are available at https://github.com/MICLab-Unicamp/medseg.
Lung2Lung: Volumetric Style Transfer with Self-Ensembling for High-Resolution Cross-Volume Computed Tomography
Oct 06, 2022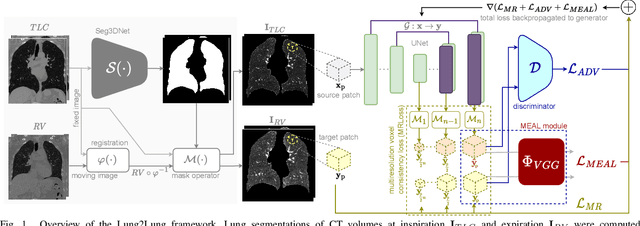
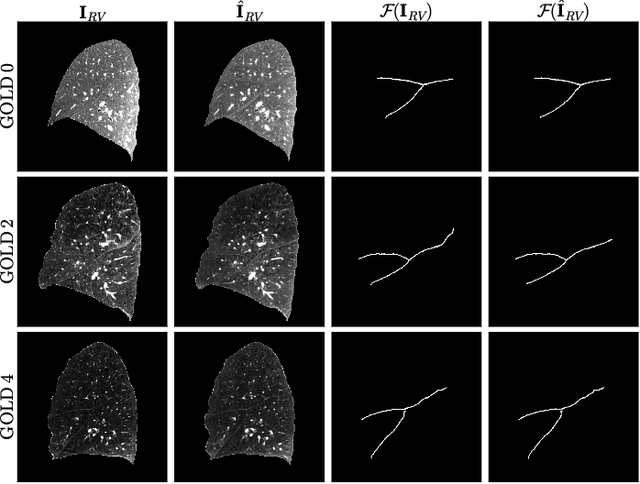
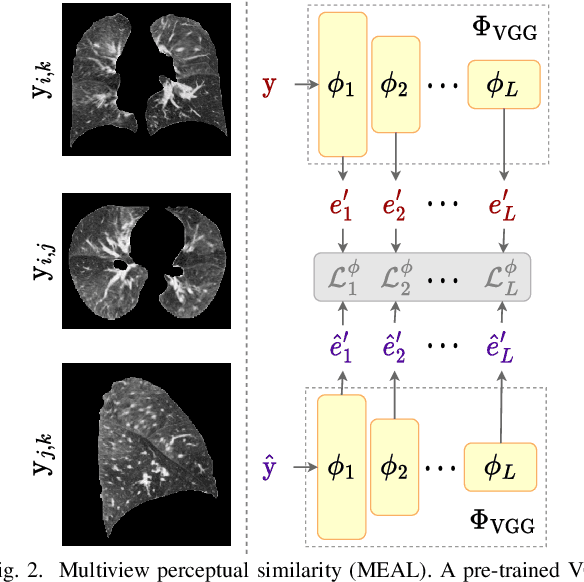
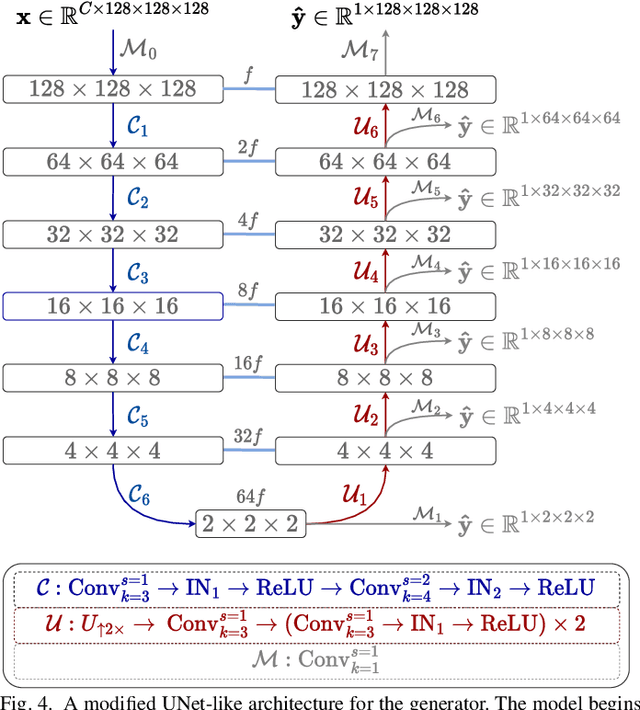
Abstract:Chest computed tomography (CT) at inspiration is often complemented by an expiratory CT for identifying peripheral airways disease in the form of air trapping. Additionally, co-registered inspiratory-expiratory volumes are used to derive several clinically relevant measures of local lung function. Acquiring CT at different volumes, however, increases radiation dosage, acquisition time, and may not be achievable due to various complications, limiting the utility of registration-based measures, To address this, we propose Lung2Lung - a style-based generative adversarial approach for translating CT images from end-inspiratory to end-expiratory volume. Lung2Lung addresses several limitations of the traditional generative models including slicewise discontinuities, limited size of generated volumes, and their inability to model neural style at a volumetric level. We introduce multiview perceptual similarity (MEAL) to capture neural styles in 3D. To incorporate global information into the training process and refine the output of our model, we also propose self-ensembling (SE). Lung2Lung, with MEAL and SE, is able to generate large 3D volumes of size 320 x 320 x 320 that are validated using a diverse cohort of 1500 subjects with varying disease severity. The model shows superior performance against several state-of-the-art 2D and 3D generative models with a peak-signal-to-noise ratio of 24.53 dB and structural similarity of 0.904. Clinical validation shows that the synthetic volumes can be used to reliably extract several clinical endpoints of chronic obstructive pulmonary disease.
Single volume lung biomechanics from chest computed tomography using a mode preserving generative adversarial network
Oct 15, 2021
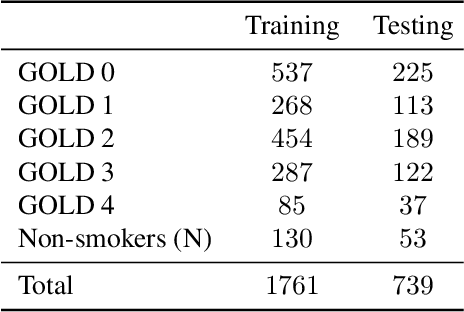
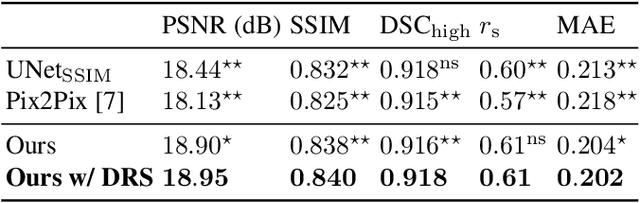
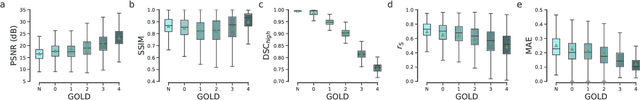
Abstract:Local tissue expansion of the lungs is typically derived by registering computed tomography (CT) scans acquired at multiple lung volumes. However, acquiring multiple scans incurs increased radiation dose, time, and cost, and may not be possible in many cases, thus restricting the applicability of registration-based biomechanics. We propose a generative adversarial learning approach for estimating local tissue expansion directly from a single CT scan. The proposed framework was trained and evaluated on 2500 subjects from the SPIROMICS cohort. Once trained, the framework can be used as a registration-free method for predicting local tissue expansion. We evaluated model performance across varying degrees of disease severity and compared its performance with two image-to-image translation frameworks - UNet and Pix2Pix. Our model achieved an overall PSNR of 18.95 decibels, SSIM of 0.840, and Spearman's correlation of 0.61 at a high spatial resolution of 1 mm3.
Geodesic Density Regression for Correcting 4DCT Pulmonary Respiratory Motion Artifacts
Jun 12, 2021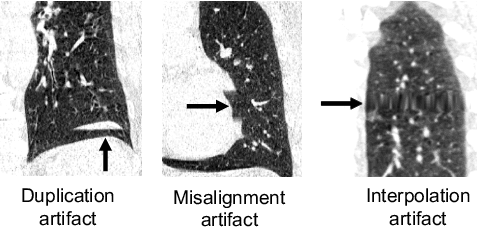
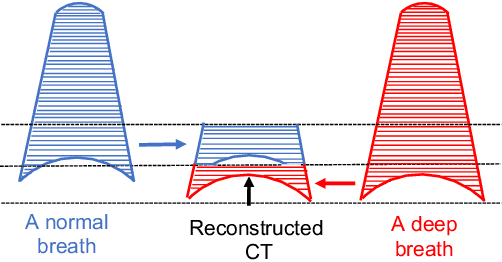
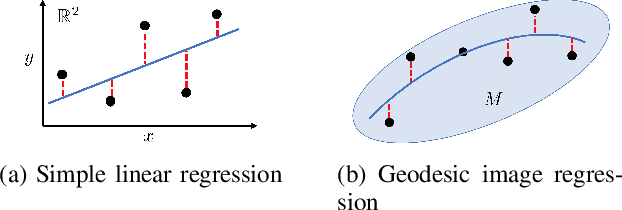
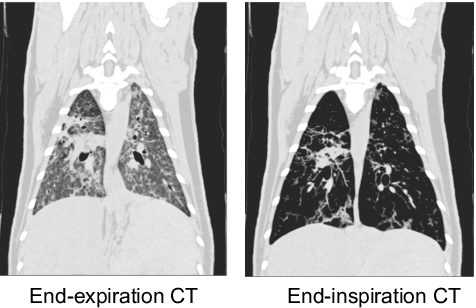
Abstract:Pulmonary respiratory motion artifacts are common in four-dimensional computed tomography (4DCT) of lungs and are caused by missing, duplicated, and misaligned image data. This paper presents a geodesic density regression (GDR) algorithm to correct motion artifacts in 4DCT by correcting artifacts in one breathing phase with artifact-free data from corresponding regions of other breathing phases. The GDR algorithm estimates an artifact-free lung template image and a smooth, dense, 4D (space plus time) vector field that deforms the template image to each breathing phase to produce an artifact-free 4DCT scan. Correspondences are estimated by accounting for the local tissue density change associated with air entering and leaving the lungs, and using binary artifact masks to exclude regions with artifacts from image regression. The artifact-free lung template image is generated by mapping the artifact-free regions of each phase volume to a common reference coordinate system using the estimated correspondences and then averaging. This procedure generates a fixed view of the lung with an improved signal-to-noise ratio. The GDR algorithm was evaluated and compared to a state-of-the-art geodesic intensity regression (GIR) algorithm using simulated CT time-series and 4DCT scans with clinically observed motion artifacts. The simulation shows that the GDR algorithm has achieved significantly more accurate Jacobian images and sharper template images, and is less sensitive to data dropout than the GIR algorithm. We also demonstrate that the GDR algorithm is more effective than the GIR algorithm for removing clinically observed motion artifacts in treatment planning 4DCT scans. Our code is freely available at https://github.com/Wei-Shao-Reg/GDR.
CT Image Segmentation for Inflamed and Fibrotic Lungs Using a Multi-Resolution Convolutional Neural Network
Oct 16, 2020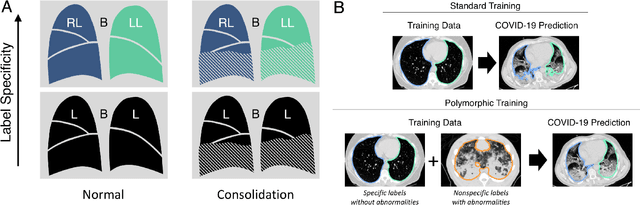
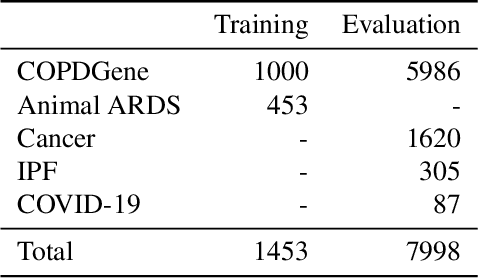
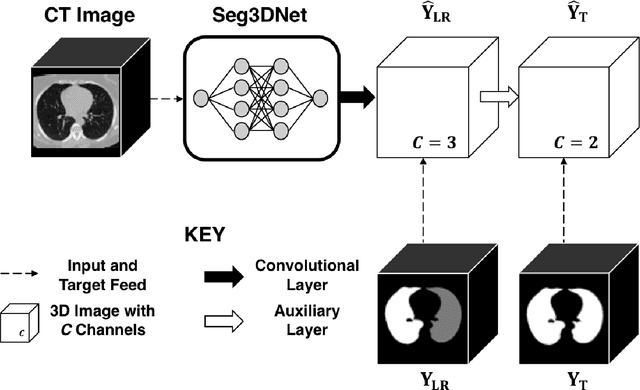
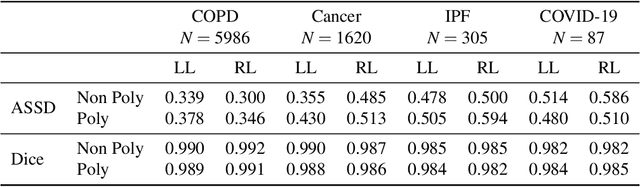
Abstract:The purpose of this study was to develop a fully-automated segmentation algorithm, robust to various density enhancing lung abnormalities, to facilitate rapid quantitative analysis of computed tomography images. A polymorphic training approach is proposed, in which both specifically labeled left and right lungs of humans with COPD, and nonspecifically labeled lungs of animals with acute lung injury, were incorporated into training a single neural network. The resulting network is intended for predicting left and right lung regions in humans with or without diffuse opacification and consolidation. Performance of the proposed lung segmentation algorithm was extensively evaluated on CT scans of subjects with COPD, confirmed COVID-19, lung cancer, and IPF, despite no labeled training data of the latter three diseases. Lobar segmentations were obtained using the left and right lung segmentation as input to the LobeNet algorithm. Regional lobar analysis was performed using hierarchical clustering to identify radiographic subtypes of COVID-19. The proposed lung segmentation algorithm was quantitatively evaluated using semi-automated and manually-corrected segmentations in 87 COVID-19 CT images, achieving an average symmetric surface distance of $0.495 \pm 0.309$ mm and Dice coefficient of $0.985 \pm 0.011$. Hierarchical clustering identified four radiographical phenotypes of COVID-19 based on lobar fractions of consolidated and poorly aerated tissue. Lower left and lower right lobes were consistently more afflicted with poor aeration and consolidation. However, the most severe cases demonstrated involvement of all lobes. The polymorphic training approach was able to accurately segment COVID-19 cases with diffuse consolidation without requiring COVID-19 cases for training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge