Muhammad F. A. Chaudhary
Uncertainty-Aware Test-Time Adaptation for Inverse Consistent Diffeomorphic Lung Image Registration
Nov 12, 2024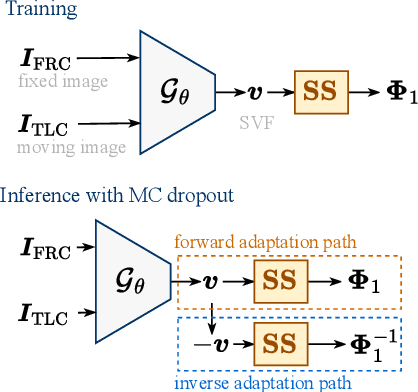
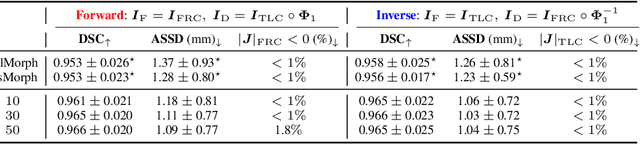
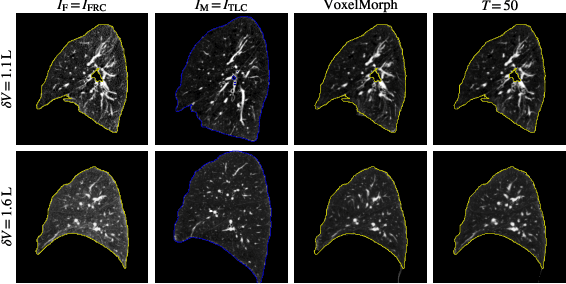
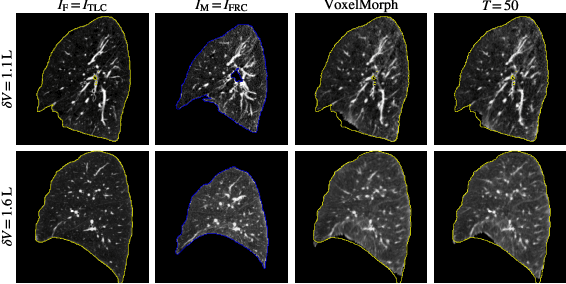
Abstract:Diffeomorphic deformable image registration ensures smooth invertible transformations across inspiratory and expiratory chest CT scans. Yet, in practice, deep learning-based diffeomorphic methods struggle to capture large deformations between inspiratory and expiratory volumes, and therefore lack inverse consistency. Existing methods also fail to account for model uncertainty, which can be useful for improving performance. We propose an uncertainty-aware test-time adaptation framework for inverse consistent diffeomorphic lung registration. Our method uses Monte Carlo (MC) dropout to estimate spatial uncertainty that is used to improve model performance. We train and evaluate our method for inspiratory-to-expiratory CT registration on a large cohort of 675 subjects from the COPDGene study, achieving a higher Dice similarity coefficient (DSC) between the lung boundaries (0.966) compared to both VoxelMorph (0.953) and TransMorph (0.953). Our method demonstrates consistent improvements in the inverse registration direction as well with an overall DSC of 0.966, higher than VoxelMorph (0.958) and TransMorph (0.956). Paired t-tests indicate statistically significant improvements.
Towards robust radiomics and radiogenomics predictive models for brain tumor characterization
Jun 05, 2024



Abstract:In the context of brain tumor characterization, we focused on two key questions: (a) stability of radiomics features to variability in multiregional segmentation masks obtained with fully-automatic deep segmentation methods and (b) subsequent impact on predictive performance on downstream tasks: IDH prediction and Overall Survival (OS) classification. We further constrained our study to limited computational resources setting which are found in underprivileged, remote, and (or) resource-starved clinical sites in developing countries. We employed seven SOTA CNNs which can be trained with limited computational resources and have demonstrated superior segmentation performance on BraTS challenge. Subsequent selection of discriminatory features was done with RFE-SVM and MRMR. Our study revealed that highly stable radiomics features were: (1) predominantly texture features (79.1%), (2) mainly extracted from WT region (96.1%), and (3) largely representing T1Gd (35.9%) and T1 (28%) sequences. Shape features and radiomics features extracted from the ENC subregion had the lowest average stability. Stability filtering minimized non-physiological variability in predictive models as indicated by an order-of-magnitude decrease in the relative standard deviation of AUCs. The non-physiological variability is attributed to variability in multiregional segmentation maps obtained with fully-automatic CNNs. Stability filtering significantly improved predictive performance on the two downstream tasks substantiating the inevitability of learning novel radiomics and radiogenomics models with stable discriminatory features. The study (implicitly) demonstrates the importance of suboptimal deep segmentation networks which can be exploited as auxiliary networks for subsequent identification of radiomics features stable to variability in automatically generated multiregional segmentation maps.
Lung2Lung: Volumetric Style Transfer with Self-Ensembling for High-Resolution Cross-Volume Computed Tomography
Oct 06, 2022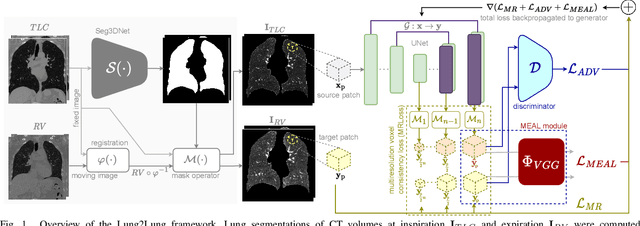
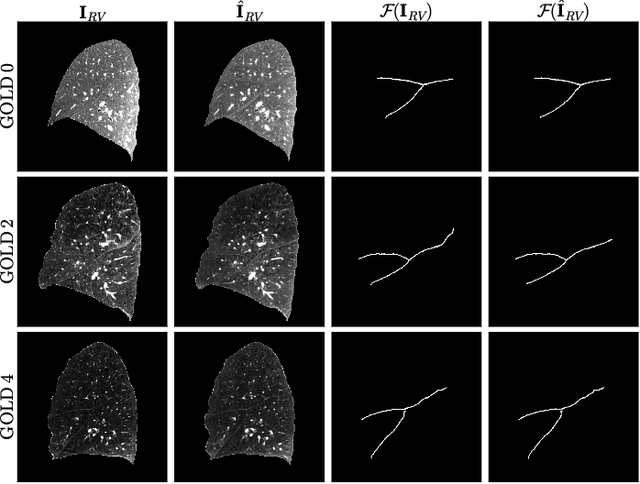
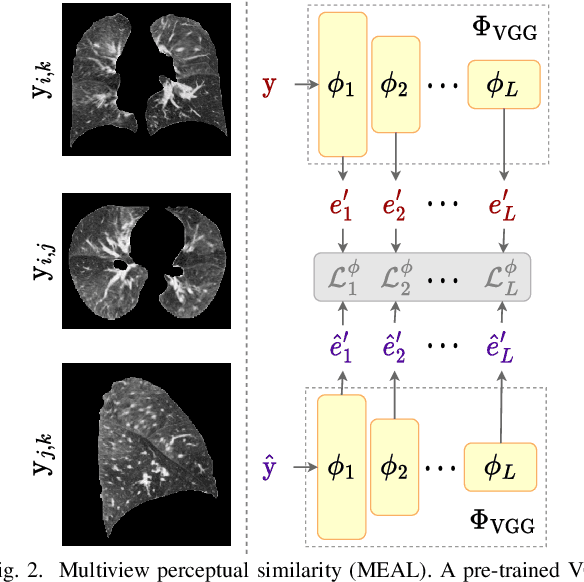
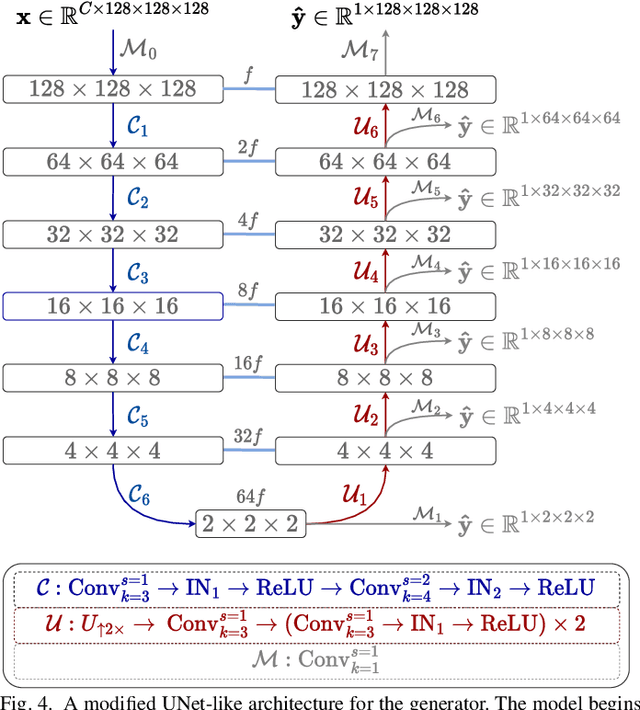
Abstract:Chest computed tomography (CT) at inspiration is often complemented by an expiratory CT for identifying peripheral airways disease in the form of air trapping. Additionally, co-registered inspiratory-expiratory volumes are used to derive several clinically relevant measures of local lung function. Acquiring CT at different volumes, however, increases radiation dosage, acquisition time, and may not be achievable due to various complications, limiting the utility of registration-based measures, To address this, we propose Lung2Lung - a style-based generative adversarial approach for translating CT images from end-inspiratory to end-expiratory volume. Lung2Lung addresses several limitations of the traditional generative models including slicewise discontinuities, limited size of generated volumes, and their inability to model neural style at a volumetric level. We introduce multiview perceptual similarity (MEAL) to capture neural styles in 3D. To incorporate global information into the training process and refine the output of our model, we also propose self-ensembling (SE). Lung2Lung, with MEAL and SE, is able to generate large 3D volumes of size 320 x 320 x 320 that are validated using a diverse cohort of 1500 subjects with varying disease severity. The model shows superior performance against several state-of-the-art 2D and 3D generative models with a peak-signal-to-noise ratio of 24.53 dB and structural similarity of 0.904. Clinical validation shows that the synthetic volumes can be used to reliably extract several clinical endpoints of chronic obstructive pulmonary disease.
Single volume lung biomechanics from chest computed tomography using a mode preserving generative adversarial network
Oct 15, 2021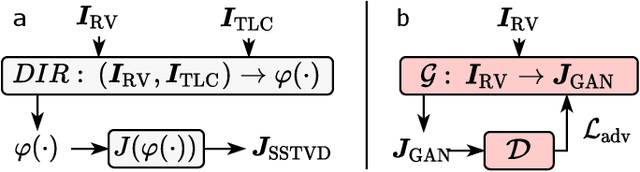
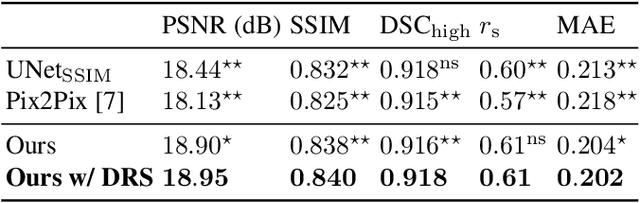
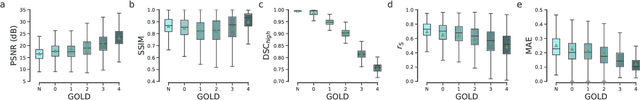
Abstract:Local tissue expansion of the lungs is typically derived by registering computed tomography (CT) scans acquired at multiple lung volumes. However, acquiring multiple scans incurs increased radiation dose, time, and cost, and may not be possible in many cases, thus restricting the applicability of registration-based biomechanics. We propose a generative adversarial learning approach for estimating local tissue expansion directly from a single CT scan. The proposed framework was trained and evaluated on 2500 subjects from the SPIROMICS cohort. Once trained, the framework can be used as a registration-free method for predicting local tissue expansion. We evaluated model performance across varying degrees of disease severity and compared its performance with two image-to-image translation frameworks - UNet and Pix2Pix. Our model achieved an overall PSNR of 18.95 decibels, SSIM of 0.840, and Spearman's correlation of 0.61 at a high spatial resolution of 1 mm3.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge