Hassan Mohy-ud-Din
Computational Imaging Meets LLMs: Zero-Shot IDH Mutation Prediction in Brain Gliomas
Nov 05, 2025Abstract:We present a framework that combines Large Language Models with computational image analytics for non-invasive, zero-shot prediction of IDH mutation status in brain gliomas. For each subject, coregistered multi-parametric MRI scans and multi-class tumor segmentation maps were processed to extract interpretable semantic (visual) attributes and quantitative features, serialized in a standardized JSON file, and used to query GPT 4o and GPT 5 without fine-tuning. We evaluated this framework on six publicly available datasets (N = 1427) and results showcased high accuracy and balanced classification performance across heterogeneous cohorts, even in the absence of manual annotations. GPT 5 outperformed GPT 4o in context-driven phenotype interpretation. Volumetric features emerged as the most important predictors, supplemented by subtype-specific imaging markers and clinical information. Our results demonstrate the potential of integrating LLM-based reasoning with computational image analytics for precise, non-invasive tumor genotyping, advancing diagnostic strategies in neuro-oncology. The code is available at https://github.com/ATPLab-LUMS/CIM-LLM.
Towards robust radiomics and radiogenomics predictive models for brain tumor characterization
Jun 05, 2024



Abstract:In the context of brain tumor characterization, we focused on two key questions: (a) stability of radiomics features to variability in multiregional segmentation masks obtained with fully-automatic deep segmentation methods and (b) subsequent impact on predictive performance on downstream tasks: IDH prediction and Overall Survival (OS) classification. We further constrained our study to limited computational resources setting which are found in underprivileged, remote, and (or) resource-starved clinical sites in developing countries. We employed seven SOTA CNNs which can be trained with limited computational resources and have demonstrated superior segmentation performance on BraTS challenge. Subsequent selection of discriminatory features was done with RFE-SVM and MRMR. Our study revealed that highly stable radiomics features were: (1) predominantly texture features (79.1%), (2) mainly extracted from WT region (96.1%), and (3) largely representing T1Gd (35.9%) and T1 (28%) sequences. Shape features and radiomics features extracted from the ENC subregion had the lowest average stability. Stability filtering minimized non-physiological variability in predictive models as indicated by an order-of-magnitude decrease in the relative standard deviation of AUCs. The non-physiological variability is attributed to variability in multiregional segmentation maps obtained with fully-automatic CNNs. Stability filtering significantly improved predictive performance on the two downstream tasks substantiating the inevitability of learning novel radiomics and radiogenomics models with stable discriminatory features. The study (implicitly) demonstrates the importance of suboptimal deep segmentation networks which can be exploited as auxiliary networks for subsequent identification of radiomics features stable to variability in automatically generated multiregional segmentation maps.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
E1D3 U-Net for Brain Tumor Segmentation: Submission to the RSNA-ASNR-MICCAI BraTS 2021 Challenge
Oct 06, 2021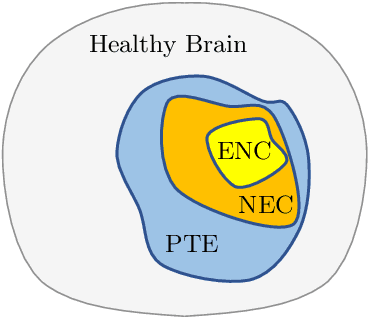
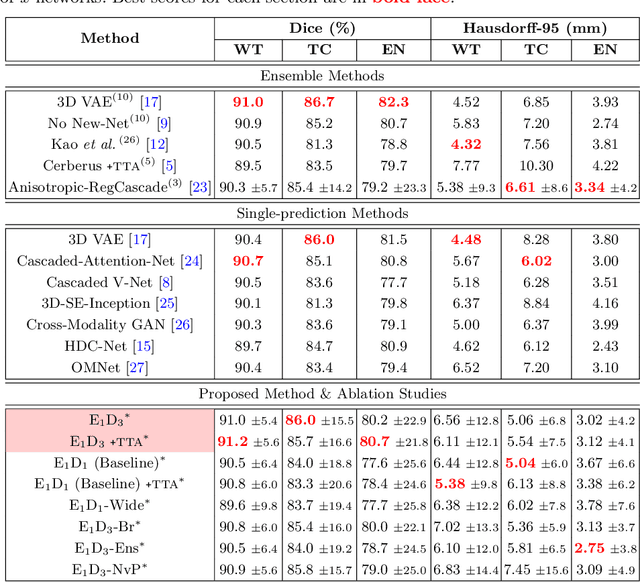
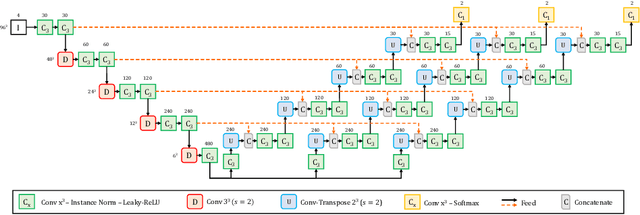
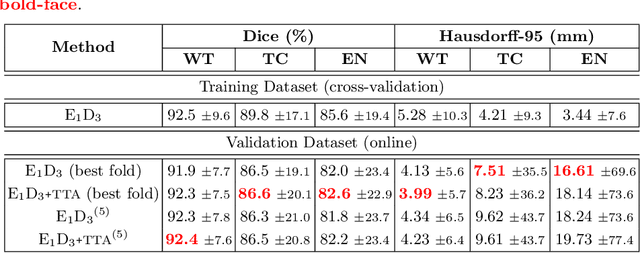
Abstract:Convolutional Neural Networks (CNNs) have demonstrated state-of-the-art performance in medical image segmentation tasks. A common feature in most top-performing CNNs is an encoder-decoder architecture inspired by the U-Net. For multi-region brain tumor segmentation, 3D U-Net architecture and its variants provide the most competitive segmentation performances. In this work, we propose an interesting extension of the standard 3D U-Net architecture, specialized for brain tumor segmentation. The proposed network, called E1D3 U-Net, is a one-encoder, three-decoder fully-convolutional neural network architecture where each decoder segments one of the hierarchical regions of interest: whole tumor, tumor core, and enhancing core. On the BraTS 2018 validation (unseen) dataset, E1D3 U-Net demonstrates single-prediction performance comparable with most state-of-the-art networks in brain tumor segmentation, with reasonable computational requirements and without ensembling. As a submission to the RSNA-ASNR-MICCAI BraTS 2021 challenge, we also evaluate our proposal on the BraTS 2021 dataset. E1D3 U-Net showcases the flexibility in the standard 3D U-Net architecture which we exploit for the task of brain tumor segmentation.
Multi-view SA-LA Net: A framework for simultaneous segmentation of RV on multi-view cardiac MR Images
Oct 01, 2021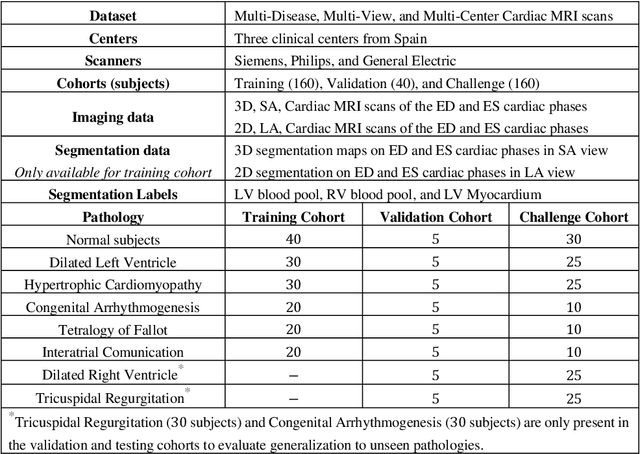

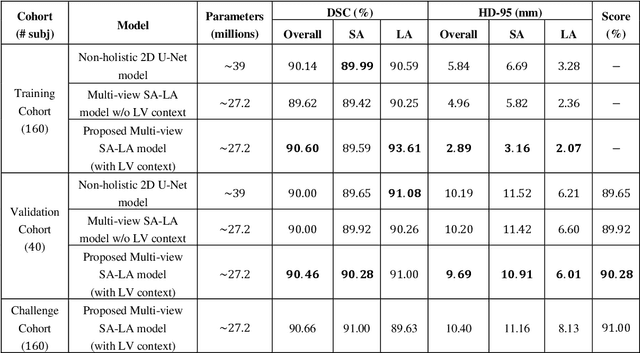
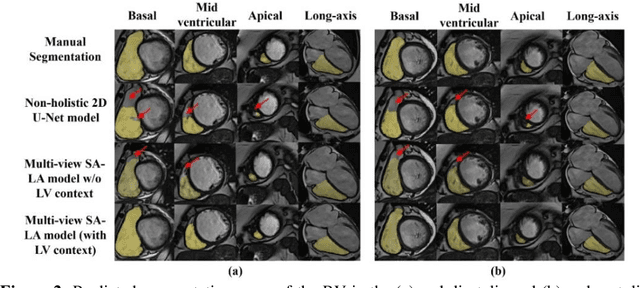
Abstract:We proposed a multi-view SA-LA model for simultaneous segmentation of RV on the short-axis (SA) and long-axis (LA) cardiac MR images. The multi-view SA-LA model is a multi-encoder, multi-decoder U-Net architecture based on the U-Net model. One encoder-decoder pair segments the RV on SA images and the other pair on LA images. Multi-view SA-LA model assembles an extremely rich set of synergistic features, at the root of the encoder branch, by combining feature maps learned from matched SA and LA cardiac MR images. Segmentation performance is further enhanced by: (1) incorporating spatial context of LV as a prior and (2) performing deep supervision in the last three layers of the decoder branch. Multi-view SA-LA model was extensively evaluated on the MICCAI 2021 Multi- Disease, Multi-View, and Multi- Centre RV Segmentation Challenge dataset (M&Ms-2021). M&Ms-2021 dataset consists of multi-phase, multi-view cardiac MR images of 360 subjects acquired at four clinical centers with three different vendors. On the challenge cohort (160 subjects), the proposed multi-view SA-LA model achieved a Dice Score of 91% and Hausdorff distance of 11.2 mm on short-axis images and a Dice Score of 89.6% and Hausdorff distance of 8.1 mm on long-axis images. Moreover, multi-view SA-LA model exhibited strong generalization to unseen RV related pathologies including Dilated Right Ventricle (DSC: SA 91.41%, LA 89.63%) and Tricuspidal Regurgitation (DSC: SA 91.40%, LA 90.40%) with low variance (std_DSC: SA <5%, LA<6%).
A Survey on Recent Advancements for AI Enabled Radiomics in Neuro-Oncology
Oct 16, 2019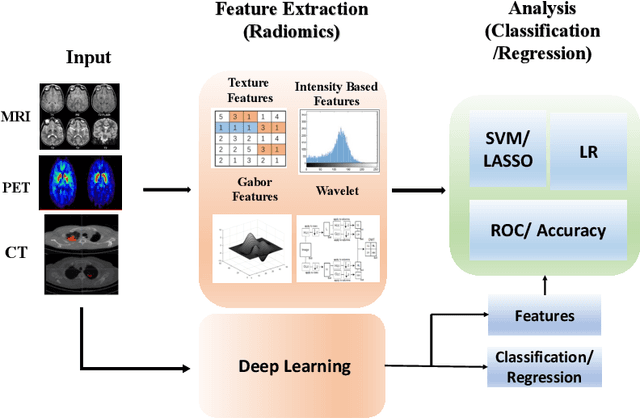
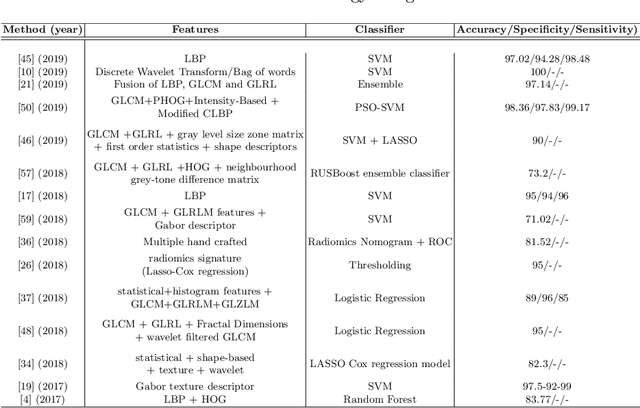
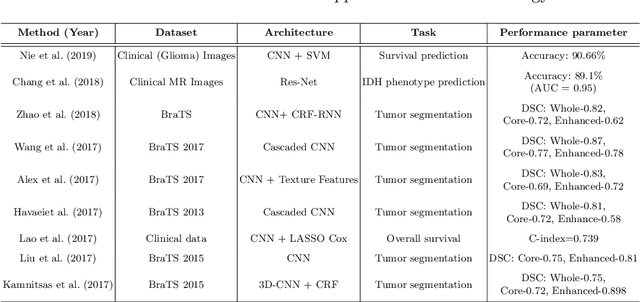
Abstract:Artificial intelligence (AI) enabled radiomics has evolved immensely especially in the field of oncology. Radiomics provide assistancein diagnosis of cancer, planning of treatment strategy, and predictionof survival. Radiomics in neuro-oncology has progressed significantly inthe recent past. Deep learning has outperformed conventional machinelearning methods in most image-based applications. Convolutional neu-ral networks (CNNs) have seen some popularity in radiomics, since theydo not require hand-crafted features and can automatically extract fea-tures during the learning process. In this regard, it is observed that CNNbased radiomics could provide state-of-the-art results in neuro-oncology,similar to the recent success of such methods in a wide spectrum ofmedical image analysis applications. Herein we present a review of the most recent best practices and establish the future trends for AI enabled radiomics in neuro-oncology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge