Ulas Bagci
Domain-invariant Mixed-domain Semi-supervised Medical Image Segmentation with Clustered Maximum Mean Discrepancy Alignment
Jan 23, 2026Abstract:Deep learning has shown remarkable progress in medical image semantic segmentation, yet its success heavily depends on large-scale expert annotations and consistent data distributions. In practice, annotations are scarce, and images are collected from multiple scanners or centers, leading to mixed-domain settings with unknown domain labels and severe domain gaps. Existing semi-supervised or domain adaptation approaches typically assume either a single domain shift or access to explicit domain indices, which rarely hold in real-world deployment. In this paper, we propose a domain-invariant mixed-domain semi-supervised segmentation framework that jointly enhances data diversity and mitigates domain bias. A Copy-Paste Mechanism (CPM) augments the training set by transferring informative regions across domains, while a Cluster Maximum Mean Discrepancy (CMMD) block clusters unlabeled features and aligns them with labeled anchors via an MMD objective, encouraging domain-invariant representations. Integrated within a teacher-student framework, our method achieves robust and precise segmentation even with very few labeled examples and multiple unknown domain discrepancies. Experiments on Fundus and M&Ms benchmarks demonstrate that our approach consistently surpasses semi-supervised and domain adaptation methods, establishing a potential solution for mixed-domain semi-supervised medical image segmentation.
CTest-Metric: A Unified Framework to Assess Clinical Validity of Metrics for CT Report Generation
Jan 16, 2026Abstract:In the generative AI era, where even critical medical tasks are increasingly automated, radiology report generation (RRG) continues to rely on suboptimal metrics for quality assessment. Developing domain-specific metrics has therefore been an active area of research, yet it remains challenging due to the lack of a unified, well-defined framework to assess their robustness and applicability in clinical contexts. To address this, we present CTest-Metric, a first unified metric assessment framework with three modules determining the clinical feasibility of metrics for CT RRG. The modules test: (i) Writing Style Generalizability (WSG) via LLM-based rephrasing; (ii) Synthetic Error Injection (SEI) at graded severities; and (iii) Metrics-vs-Expert correlation (MvE) using clinician ratings on 175 "disagreement" cases. Eight widely used metrics (BLEU, ROUGE, METEOR, BERTScore-F1, F1-RadGraph, RaTEScore, GREEN Score, CRG) are studied across seven LLMs built on a CT-CLIP encoder. Using our novel framework, we found that lexical NLG metrics are highly sensitive to stylistic variations; GREEN Score aligns best with expert judgments (Spearman~0.70), while CRG shows negative correlation; and BERTScore-F1 is least sensitive to factual error injection. We will release the framework, code, and allowable portion of the anonymized evaluation data (rephrased/error-injected CT reports), to facilitate reproducible benchmarking and future metric development.
Ensemble Models for Predicting Treatment Response in Pediatric Low-Grade Glioma Managed with Chemotherapy
Jan 07, 2026Abstract:In this paper, we introduce a novel pipeline for predicting chemotherapy response in pediatric brain tumors that are not amenable to complete surgical resection, using pre-treatment magnetic resonance imaging combined with clinical information. Our method integrates a state-of-the-art pediatric brain tumor segmentation framework with radiomic feature extraction and clinical data through an ensemble of a Swin UNETR encoder and XGBoost classifier. The segmentation model delineates four tumor subregions enhancing tumor, non-enhancing tumor, cystic component and edema which are used to extract imaging biomarkers and generate predictive features. The Swin UNETR network classifies the response to treatment directly from these segmented MRI scans, while XGBoost predicts response using radiomics and clinical variables including legal sex, ethnicity, race, age at event (in days), molecular subtype, tumor locations, initial surgery status, metastatic status, metastasis location, chemotherapy type, protocol name and chemotherapy agents. The ensemble output provides a non-invasive estimate of chemotherapy response in this historically challenging population characterized by lower progression-free survival. Among compared approaches, our Swin-Ensemble achieved the best performance (precision for non effective cases=0.68, recall for non effective cases=0.85, precision for chemotherapy effective cases=0.64 and overall accuracy=0.69), outperforming Mamba-FeatureFuse, Swin UNETR encoder, and Swin-FeatureFuse models. Our findings suggest that this ensemble framework represents a promising step toward personalized therapy response prediction for pediatric low-grade glioma patients in need of chemotherapy treatment who are not suitable for complete surgical resection, a population with significantly lower progression free survival and for whom chemotherapy remains the primary treatment option.
ProDM: Synthetic Reality-driven Property-aware Progressive Diffusion Model for Coronary Calcium Motion Correction in Non-gated Chest CT
Dec 31, 2025Abstract:Coronary artery calcium (CAC) scoring from chest CT is a well-established tool to stratify and refine clinical cardiovascular disease risk estimation. CAC quantification relies on the accurate delineation of calcified lesions, but is oftentimes affected by artifacts introduced by cardiac and respiratory motion. ECG-gated cardiac CTs substantially reduce motion artifacts, but their use in population screening and routine imaging remains limited due to gating requirements and lack of insurance coverage. Although identification of incidental CAC from non-gated chest CT is increasingly considered for it offers an accessible and widely available alternative, this modality is limited by more severe motion artifacts. We present ProDM (Property-aware Progressive Correction Diffusion Model), a generative diffusion framework that restores motion-free calcified lesions from non-gated CTs. ProDM introduces three key components: (1) a CAC motion simulation data engine that synthesizes realistic non-gated acquisitions with diverse motion trajectories directly from cardiac-gated CTs, enabling supervised training without paired data; (2) a property-aware learning strategy incorporating calcium-specific priors through a differentiable calcium consistency loss to preserve lesion integrity; and (3) a progressive correction scheme that reduces artifacts gradually across diffusion steps to enhance stability and calcium fidelity. Experiments on real patient datasets show that ProDM significantly improves CAC scoring accuracy, spatial lesion fidelity, and risk stratification performance compared with several baselines. A reader study on real non-gated scans further confirms that ProDM suppresses motion artifacts and improves clinical usability. These findings highlight the potential of progressive, property-aware frameworks for reliable CAC quantification from routine chest CT imaging.
REN: Anatomically-Informed Mixture-of-Experts for Interstitial Lung Disease Diagnosis
Oct 06, 2025Abstract:Mixture-of-Experts (MoE) architectures have significantly contributed to scalable machine learning by enabling specialized subnetworks to tackle complex tasks efficiently. However, traditional MoE systems lack domain-specific constraints essential for medical imaging, where anatomical structure and regional disease heterogeneity strongly influence pathological patterns. Here, we introduce Regional Expert Networks (REN), the first anatomically-informed MoE framework tailored specifically for medical image classification. REN leverages anatomical priors to train seven specialized experts, each dedicated to distinct lung lobes and bilateral lung combinations, enabling precise modeling of region-specific pathological variations. Multi-modal gating mechanisms dynamically integrate radiomics biomarkers and deep learning (DL) features (CNN, ViT, Mamba) to weight expert contributions optimally. Applied to interstitial lung disease (ILD) classification, REN achieves consistently superior performance: the radiomics-guided ensemble reached an average AUC of 0.8646 +/- 0.0467, a +12.5 percent improvement over the SwinUNETR baseline (AUC 0.7685, p = 0.031). Region-specific experts further revealed that lower-lobe models achieved AUCs of 0.88-0.90, surpassing DL counterparts (CNN: 0.76-0.79) and aligning with known disease progression patterns. Through rigorous patient-level cross-validation, REN demonstrates strong generalizability and clinical interpretability, presenting a scalable, anatomically-guided approach readily extensible to other structured medical imaging applications.
Label-Efficient Cross-Modality Generalization for Liver Segmentation in Multi-Phase MRI
Oct 06, 2025Abstract:Accurate liver segmentation in multi-phase MRI is vital for liver fibrosis assessment, yet labeled data is often scarce and unevenly distributed across imaging modalities and vendor systems. We propose a label-efficient segmentation approach that promotes cross-modality generalization under real-world conditions, where GED4 hepatobiliary-phase annotations are limited, non-contrast sequences (T1WI, T2WI, DWI) are unlabeled, and spatial misalignment and missing phases are common. Our method integrates a foundation-scale 3D segmentation backbone adapted via fine-tuning, co-training with cross pseudo supervision to leverage unlabeled volumes, and a standardized preprocessing pipeline. Without requiring spatial registration, the model learns to generalize across MRI phases and vendors, demonstrating robust segmentation performance in both labeled and unlabeled domains. Our results exhibit the effectiveness of our proposed label-efficient baseline for liver segmentation in multi-phase, multi-vendor MRI and highlight the potential of combining foundation model adaptation with co-training for real-world clinical imaging tasks.
SRMA-Mamba: Spatial Reverse Mamba Attention Network for Pathological Liver Segmentation in MRI Volumes
Aug 17, 2025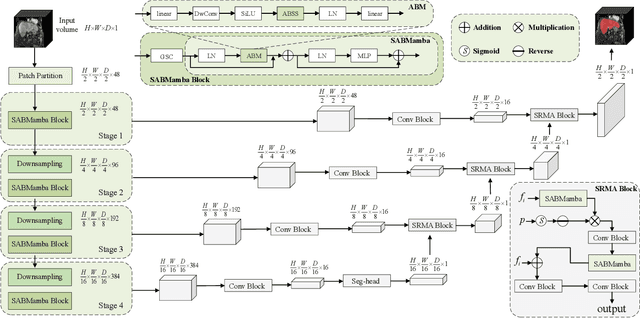
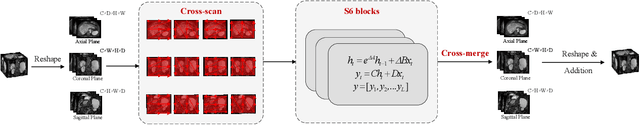
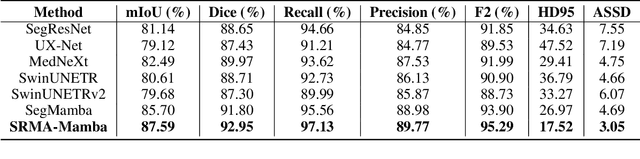
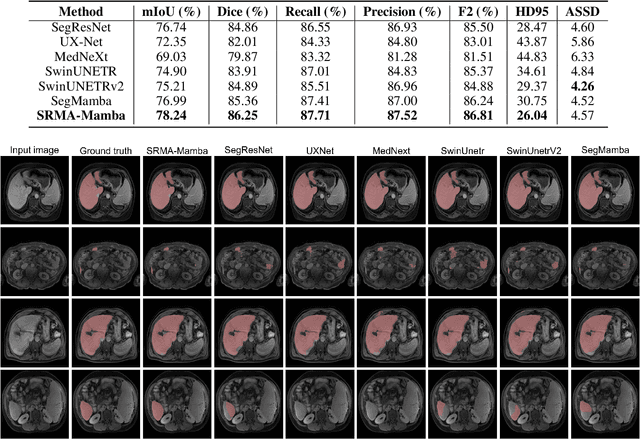
Abstract:Liver Cirrhosis plays a critical role in the prognosis of chronic liver disease. Early detection and timely intervention are critical in significantly reducing mortality rates. However, the intricate anatomical architecture and diverse pathological changes of liver tissue complicate the accurate detection and characterization of lesions in clinical settings. Existing methods underutilize the spatial anatomical details in volumetric MRI data, thereby hindering their clinical effectiveness and explainability. To address this challenge, we introduce a novel Mamba-based network, SRMA-Mamba, designed to model the spatial relationships within the complex anatomical structures of MRI volumes. By integrating the Spatial Anatomy-Based Mamba module (SABMamba), SRMA-Mamba performs selective Mamba scans within liver cirrhotic tissues and combines anatomical information from the sagittal, coronal, and axial planes to construct a global spatial context representation, enabling efficient volumetric segmentation of pathological liver structures. Furthermore, we introduce the Spatial Reverse Attention module (SRMA), designed to progressively refine cirrhotic details in the segmentation map, utilizing both the coarse segmentation map and hierarchical encoding features. Extensive experiments demonstrate that SRMA-Mamba surpasses state-of-the-art methods, delivering exceptional performance in 3D pathological liver segmentation. Our code is available for public: {\color{blue}{https://github.com/JunZengz/SRMA-Mamba}}.
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025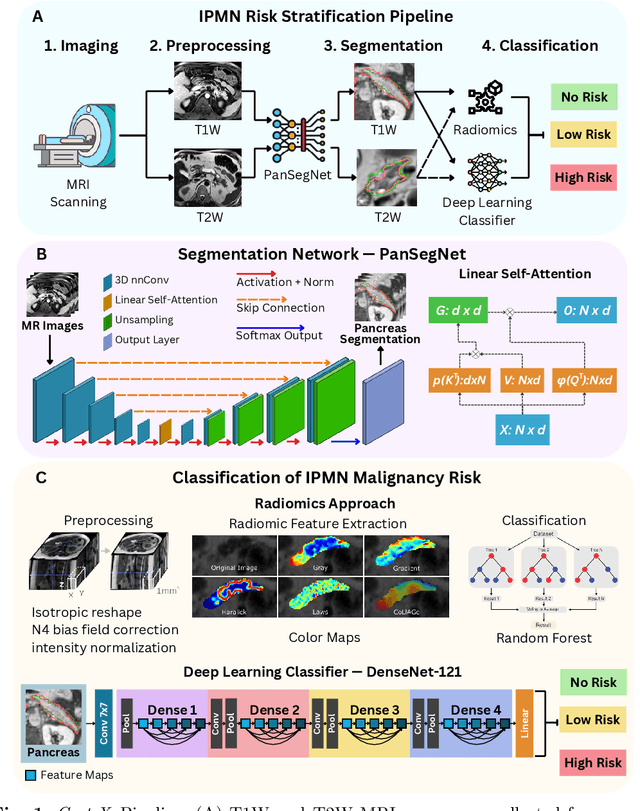

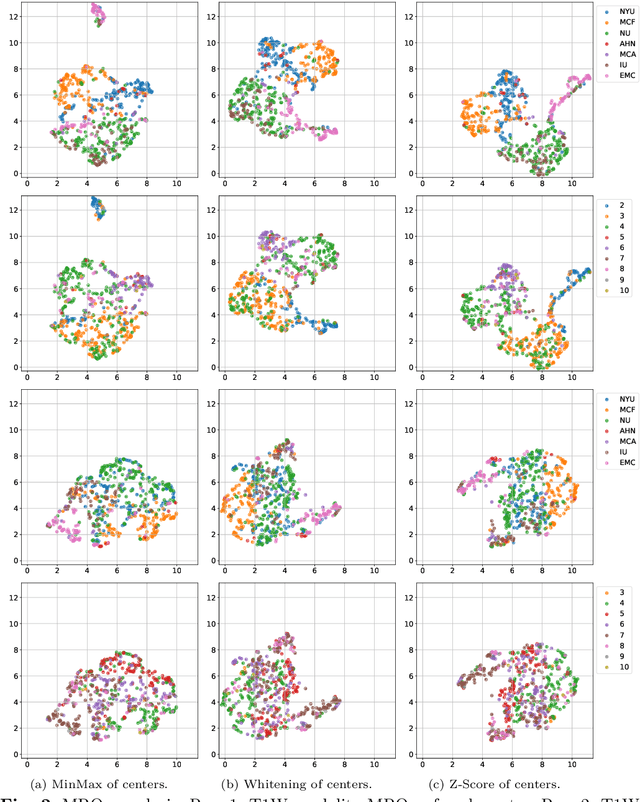
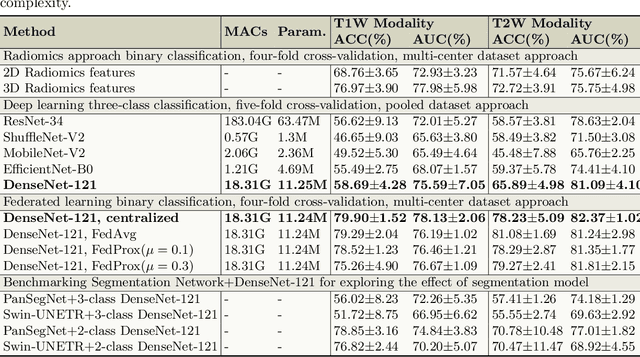
Abstract:Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
Describe Anything Model for Visual Question Answering on Text-rich Images
Jul 16, 2025Abstract:Recent progress has been made in region-aware vision-language modeling, particularly with the emergence of the Describe Anything Model (DAM). DAM is capable of generating detailed descriptions of any specific image areas or objects without the need for additional localized image-text alignment supervision. We hypothesize that such region-level descriptive capability is beneficial for the task of Visual Question Answering (VQA), especially in challenging scenarios involving images with dense text. In such settings, the fine-grained extraction of textual information is crucial to producing correct answers. Motivated by this, we introduce DAM-QA, a framework with a tailored evaluation protocol, developed to investigate and harness the region-aware capabilities from DAM for the text-rich VQA problem that requires reasoning over text-based information within images. DAM-QA incorporates a mechanism that aggregates answers from multiple regional views of image content, enabling more effective identification of evidence that may be tied to text-related elements. Experiments on six VQA benchmarks show that our approach consistently outperforms the baseline DAM, with a notable 7+ point gain on DocVQA. DAM-QA also achieves the best overall performance among region-aware models with fewer parameters, significantly narrowing the gap with strong generalist VLMs. These results highlight the potential of DAM-like models for text-rich and broader VQA tasks when paired with efficient usage and integration strategies. Our code is publicly available at https://github.com/Linvyl/DAM-QA.git.
TAGS: 3D Tumor-Adaptive Guidance for SAM
May 21, 2025Abstract:Foundation models (FMs) such as CLIP and SAM have recently shown great promise in image segmentation tasks, yet their adaptation to 3D medical imaging-particularly for pathology detection and segmentation-remains underexplored. A critical challenge arises from the domain gap between natural images and medical volumes: existing FMs, pre-trained on 2D data, struggle to capture 3D anatomical context, limiting their utility in clinical applications like tumor segmentation. To address this, we propose an adaptation framework called TAGS: Tumor Adaptive Guidance for SAM, which unlocks 2D FMs for 3D medical tasks through multi-prompt fusion. By preserving most of the pre-trained weights, our approach enhances SAM's spatial feature extraction using CLIP's semantic insights and anatomy-specific prompts. Extensive experiments on three open-source tumor segmentation datasets prove that our model surpasses the state-of-the-art medical image segmentation models (+46.88% over nnUNet), interactive segmentation frameworks, and other established medical FMs, including SAM-Med2D, SAM-Med3D, SegVol, Universal, 3D-Adapter, and SAM-B (at least +13% over them). This highlights the robustness and adaptability of our proposed framework across diverse medical segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge