Hongyi Pan
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025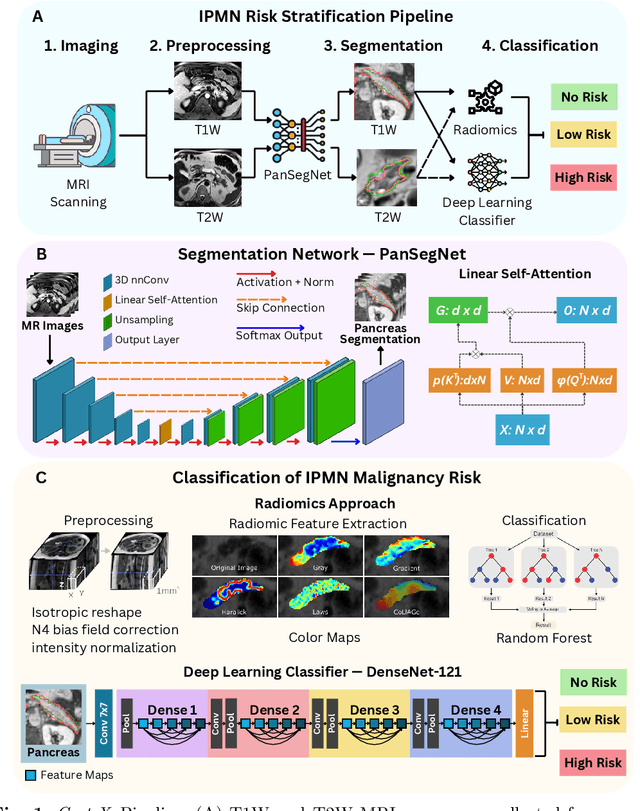

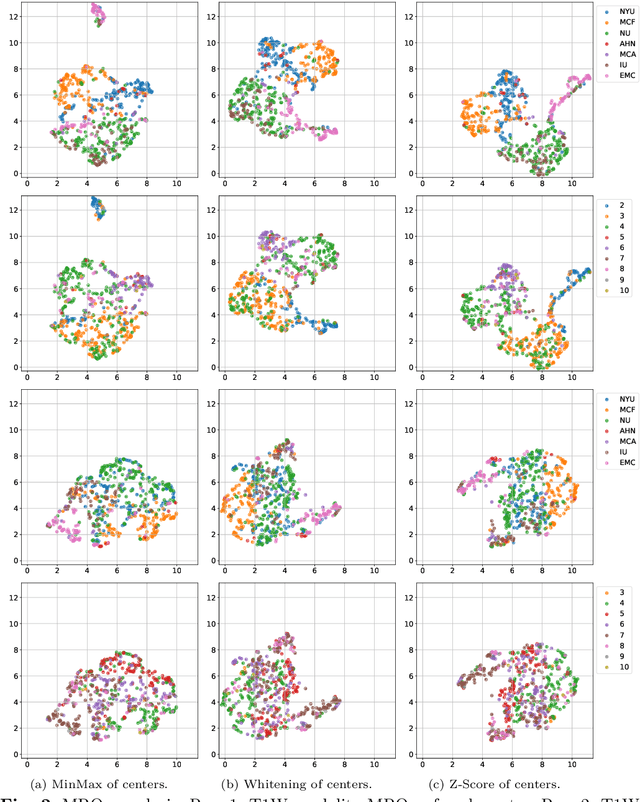
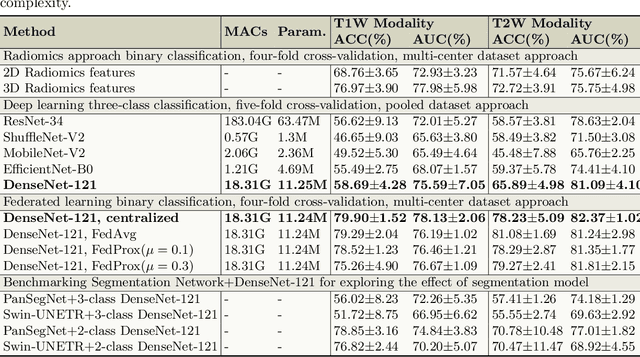
Abstract:Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
VideoAds for Fast-Paced Video Understanding: Where Opensource Foundation Models Beat GPT-4o & Gemini-1.5 Pro
Apr 12, 2025Abstract:Advertisement videos serve as a rich and valuable source of purpose-driven information, encompassing high-quality visual, textual, and contextual cues designed to engage viewers. They are often more complex than general videos of similar duration due to their structured narratives and rapid scene transitions, posing significant challenges to multi-modal large language models (MLLMs). In this work, we introduce VideoAds, the first dataset tailored for benchmarking the performance of MLLMs on advertisement videos. VideoAds comprises well-curated advertisement videos with complex temporal structures, accompanied by \textbf{manually} annotated diverse questions across three core tasks: visual finding, video summary, and visual reasoning. We propose a quantitative measure to compare VideoAds against existing benchmarks in terms of video complexity. Through extensive experiments, we find that Qwen2.5-VL-72B, an opensource MLLM, achieves 73.35\% accuracy on VideoAds, outperforming GPT-4o (66.82\%) and Gemini-1.5 Pro (69.66\%); the two proprietary models especially fall behind the opensource model in video summarization and reasoning, but perform the best in visual finding. Notably, human experts easily achieve a remarkable accuracy of 94.27\%. These results underscore the necessity of advancing MLLMs' temporal modeling capabilities and highlight VideoAds as a potentially pivotal benchmark for future research in understanding video that requires high FPS sampling. The dataset and evaluation code will be publicly available at https://videoadsbenchmark.netlify.app.
Edge-Fog Computing-Enabled EEG Data Compression via Asymmetrical Variational Discrete Cosine Transform Network
Mar 13, 2025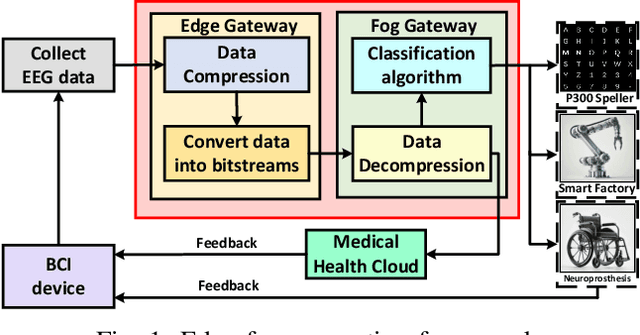
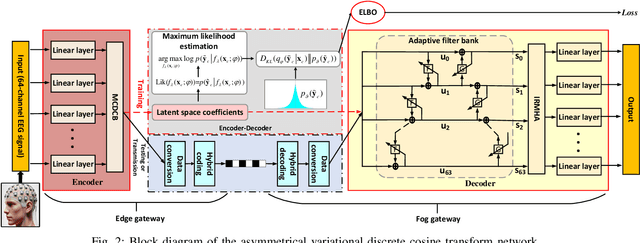
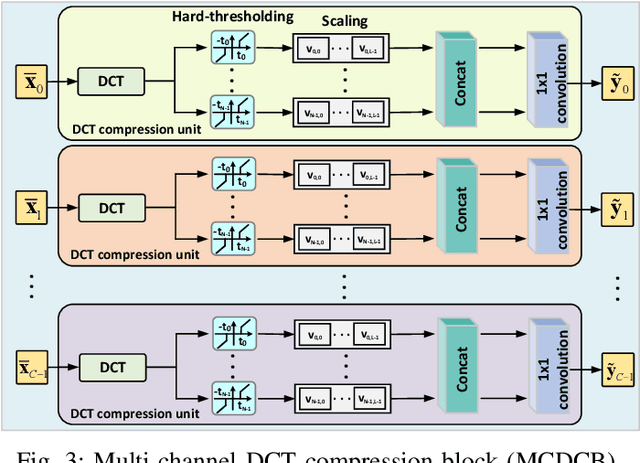
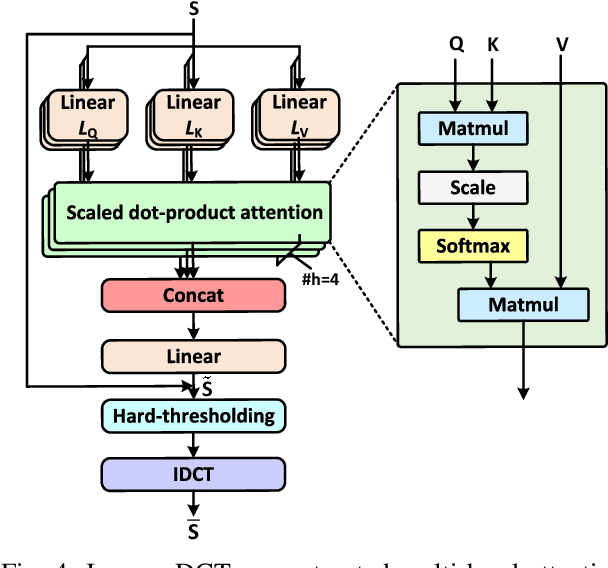
Abstract:The large volume of electroencephalograph (EEG) data produced by brain-computer interface (BCI) systems presents challenges for rapid transmission over bandwidth-limited channels in Internet of Things (IoT) networks. To address the issue, we propose a novel multi-channel asymmetrical variational discrete cosine transform (DCT) network for EEG data compression within an edge-fog computing framework. At the edge level, low-complexity DCT compression units are designed using parallel trainable hard-thresholding and scaling operators to remove redundant data and extract the effective latent space representation. At the fog level, an adaptive filter bank is applied to merge important features from adjacent channels into each individual channel by leveraging inter-channel correlations. Then, the inverse DCT reconstructed multi-head attention is developed to capture both local and global dependencies and reconstruct the original signals. Furthermore, by applying the principles of variational inference, a new evidence lower bound is formulated as the loss function, driving the model to balance compression efficiency and reconstruction accuracy. Experimental results on two public datasets demonstrate that the proposed method achieves superior compression performance without sacrificing any useful information for BCI detection compared with state-of-the-art techniques, indicating a feasible solution for EEG data compression.
Classroom Simulacra: Building Contextual Student Generative Agents in Online Education for Learning Behavioral Simulation
Feb 04, 2025



Abstract:Student simulation supports educators to improve teaching by interacting with virtual students. However, most existing approaches ignore the modulation effects of course materials because of two challenges: the lack of datasets with granularly annotated course materials, and the limitation of existing simulation models in processing extremely long textual data. To solve the challenges, we first run a 6-week education workshop from N = 60 students to collect fine-grained data using a custom built online education system, which logs students' learning behaviors as they interact with lecture materials over time. Second, we propose a transferable iterative reflection (TIR) module that augments both prompting-based and finetuning-based large language models (LLMs) for simulating learning behaviors. Our comprehensive experiments show that TIR enables the LLMs to perform more accurate student simulation than classical deep learning models, even with limited demonstration data. Our TIR approach better captures the granular dynamism of learning performance and inter-student correlations in classrooms, paving the way towards a ''digital twin'' for online education.
IPMN Risk Assessment under Federated Learning Paradigm
Nov 08, 2024



Abstract:Accurate classification of Intraductal Papillary Mucinous Neoplasms (IPMN) is essential for identifying high-risk cases that require timely intervention. In this study, we develop a federated learning framework for multi-center IPMN classification utilizing a comprehensive pancreas MRI dataset. This dataset includes 653 T1-weighted and 656 T2-weighted MRI images, accompanied by corresponding IPMN risk scores from 7 leading medical institutions, making it the largest and most diverse dataset for IPMN classification to date. We assess the performance of DenseNet-121 in both centralized and federated settings for training on distributed data. Our results demonstrate that the federated learning approach achieves high classification accuracy comparable to centralized learning while ensuring data privacy across institutions. This work marks a significant advancement in collaborative IPMN classification, facilitating secure and high-accuracy model training across multiple centers.
Adaptive Aggregation Weights for Federated Segmentation of Pancreas MRI
Oct 29, 2024

Abstract:Federated learning (FL) enables collaborative model training across institutions without sharing sensitive data, making it an attractive solution for medical imaging tasks. However, traditional FL methods, such as Federated Averaging (FedAvg), face difficulties in generalizing across domains due to variations in imaging protocols and patient demographics across institutions. This challenge is particularly evident in pancreas MRI segmentation, where anatomical variability and imaging artifacts significantly impact performance. In this paper, we conduct a comprehensive evaluation of FL algorithms for pancreas MRI segmentation and introduce a novel approach that incorporates adaptive aggregation weights. By dynamically adjusting the contribution of each client during model aggregation, our method accounts for domain-specific differences and improves generalization across heterogeneous datasets. Experimental results demonstrate that our approach enhances segmentation accuracy and reduces the impact of domain shift compared to conventional FL methods while maintaining privacy-preserving capabilities. Significant performance improvements are observed across multiple hospitals (centers).
Frequency-Based Federated Domain Generalization for Polyp Segmentation
Oct 02, 2024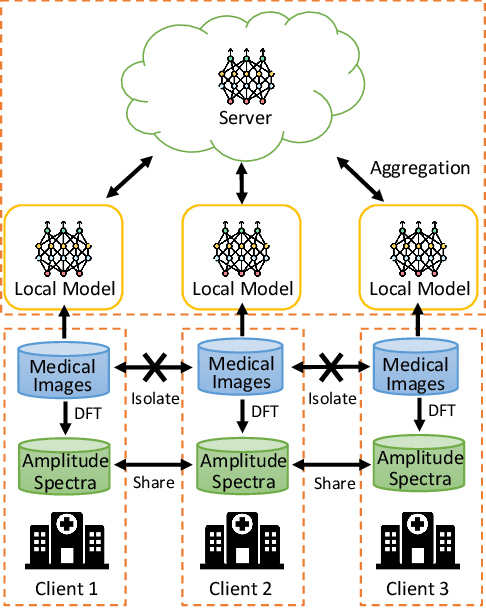
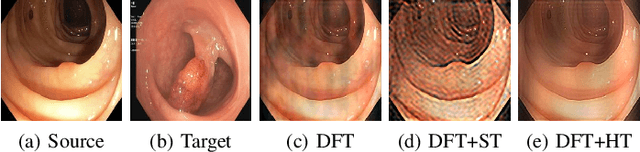
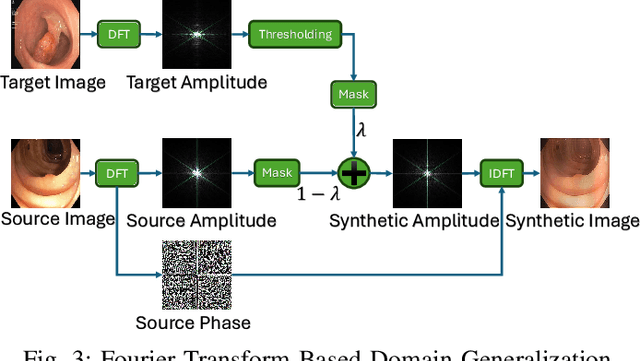

Abstract:Federated Learning (FL) offers a powerful strategy for training machine learning models across decentralized datasets while maintaining data privacy, yet domain shifts among clients can degrade performance, particularly in medical imaging tasks like polyp segmentation. This paper introduces a novel Frequency-Based Domain Generalization (FDG) framework, utilizing soft- and hard-thresholding in the Fourier domain to address these challenges. By applying soft- and hard-thresholding to Fourier coefficients, our method generates new images with reduced background noise and enhances the model's ability to generalize across diverse medical imaging domains. Extensive experiments demonstrate substantial improvements in segmentation accuracy and domain robustness over baseline methods. This innovation integrates frequency domain techniques into FL, presenting a resilient approach to overcoming domain variability in decentralized medical image analysis.
Sparse Mamba: Reinforcing Controllability In Structural State Space Models
Aug 31, 2024

Abstract:In this article, we introduce the concept of controllability and observability to the M amba architecture in our Sparse-Mamba (S-Mamba) for natural language processing (NLP) applications. The structured state space model (SSM) development in recent studies, such as Mamba and Mamba2, outperformed and solved the computational inefficiency of transformers and large language models (LLMs) on longer sequences in small to medium NLP tasks. The Mamba SSMs architecture drops the need for attention layer or MLB blocks in transformers. However, the current Mamba models do not reinforce the controllability on state space equations in the calculation of A, B, C, and D matrices at each time step, which increase the complexity and the computational cost needed. In this article we show that the number of parameters can be significantly decreased by reinforcing controllability in the state space equations in the proposed Sparse-Mamba (S-Mamba), while maintaining the performance. The controllable n x n state matrix A is sparse and it has only n free parameters. Our novel approach will ensure a controllable system and could be the gate key for Mamba 3.
DCT-Based Decorrelated Attention for Vision Transformers
May 22, 2024



Abstract:Central to the Transformer architectures' effectiveness is the self-attention mechanism, a function that maps queries, keys, and values into a high-dimensional vector space. However, training the attention weights of queries, keys, and values is non-trivial from a state of random initialization. In this paper, we propose two methods. (i) We first address the initialization problem of Vision Transformers by introducing a simple, yet highly innovative, initialization approach utilizing Discrete Cosine Transform (DCT) coefficients. Our proposed DCT-based attention initialization marks a significant gain compared to traditional initialization strategies; offering a robust foundation for the attention mechanism. Our experiments reveal that the DCT-based initialization enhances the accuracy of Vision Transformers in classification tasks. (ii) We also recognize that since DCT effectively decorrelates image information in the frequency domain, this decorrelation is useful for compression because it allows the quantization step to discard many of the higher-frequency components. Based on this observation, we propose a novel DCT-based compression technique for the attention function of Vision Transformers. Since high-frequency DCT coefficients usually correspond to noise, we truncate the high-frequency DCT components of the input patches. Our DCT-based compression reduces the size of weight matrices for queries, keys, and values. While maintaining the same level of accuracy, our DCT compressed Swin Transformers obtain a considerable decrease in the computational overhead.
Large-Scale Multi-Center CT and MRI Segmentation of Pancreas with Deep Learning
May 20, 2024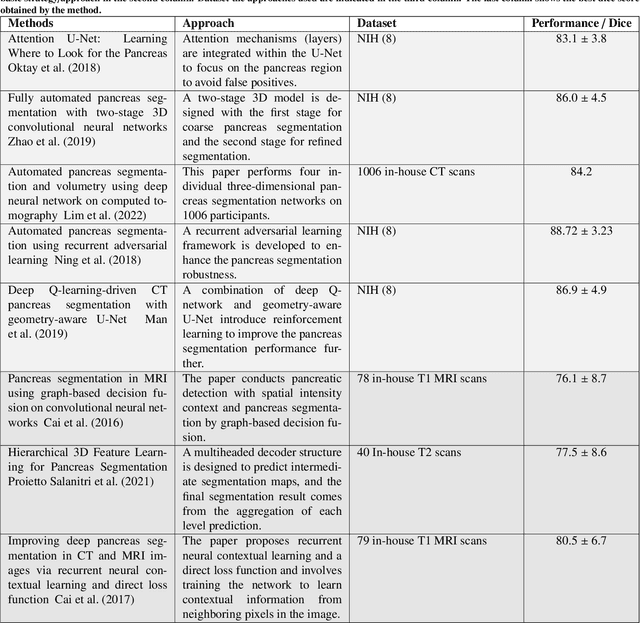
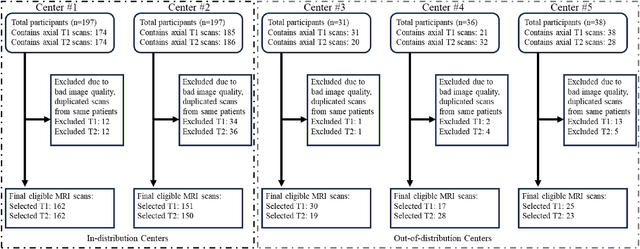
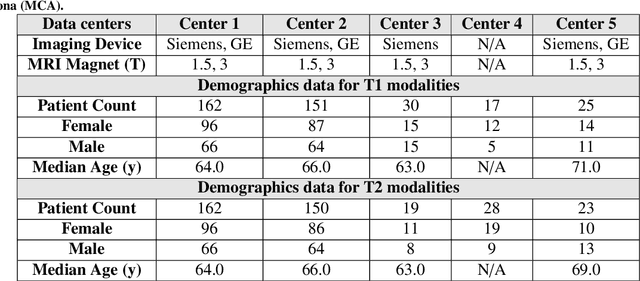
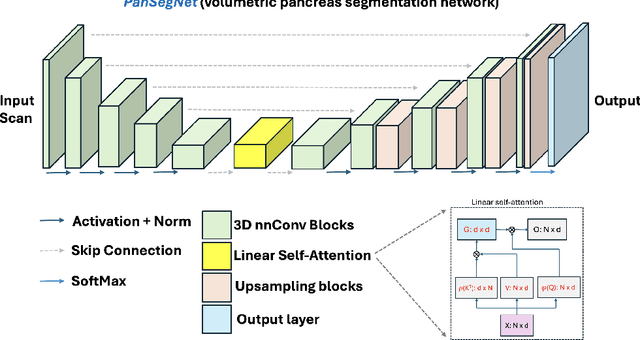
Abstract:Automated volumetric segmentation of the pancreas on cross-sectional imaging is needed for diagnosis and follow-up of pancreatic diseases. While CT-based pancreatic segmentation is more established, MRI-based segmentation methods are understudied, largely due to a lack of publicly available datasets, benchmarking research efforts, and domain-specific deep learning methods. In this retrospective study, we collected a large dataset (767 scans from 499 participants) of T1-weighted (T1W) and T2-weighted (T2W) abdominal MRI series from five centers between March 2004 and November 2022. We also collected CT scans of 1,350 patients from publicly available sources for benchmarking purposes. We developed a new pancreas segmentation method, called PanSegNet, combining the strengths of nnUNet and a Transformer network with a new linear attention module enabling volumetric computation. We tested PanSegNet's accuracy in cross-modality (a total of 2,117 scans) and cross-center settings with Dice and Hausdorff distance (HD95) evaluation metrics. We used Cohen's kappa statistics for intra and inter-rater agreement evaluation and paired t-tests for volume and Dice comparisons, respectively. For segmentation accuracy, we achieved Dice coefficients of 88.3% (std: 7.2%, at case level) with CT, 85.0% (std: 7.9%) with T1W MRI, and 86.3% (std: 6.4%) with T2W MRI. There was a high correlation for pancreas volume prediction with R^2 of 0.91, 0.84, and 0.85 for CT, T1W, and T2W, respectively. We found moderate inter-observer (0.624 and 0.638 for T1W and T2W MRI, respectively) and high intra-observer agreement scores. All MRI data is made available at https://osf.io/kysnj/. Our source code is available at https://github.com/NUBagciLab/PaNSegNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge