Yavuz Taktak
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025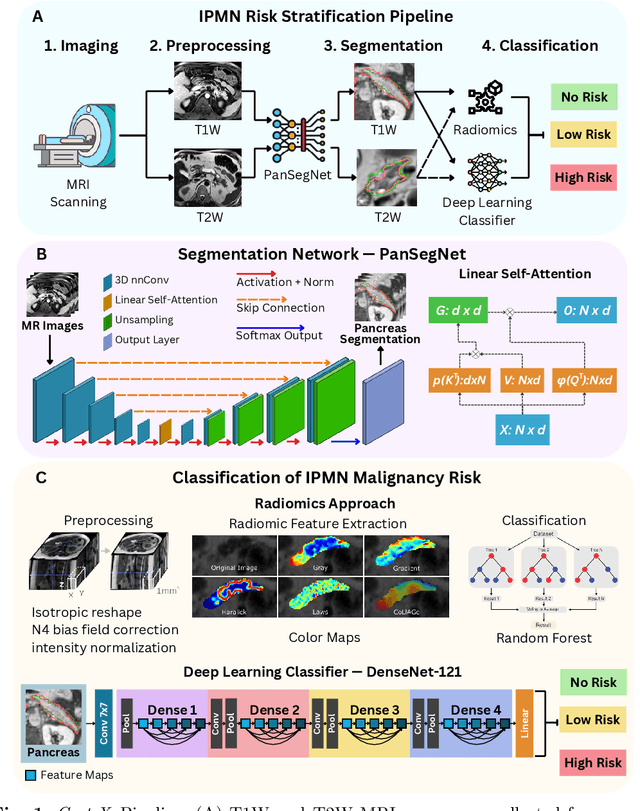

Abstract:Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
IPMN Risk Assessment under Federated Learning Paradigm
Nov 08, 2024



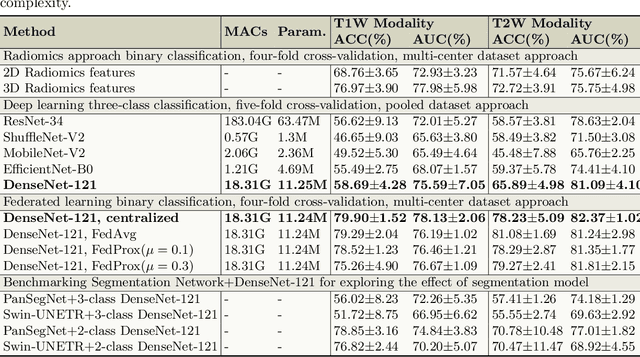
Abstract:Accurate classification of Intraductal Papillary Mucinous Neoplasms (IPMN) is essential for identifying high-risk cases that require timely intervention. In this study, we develop a federated learning framework for multi-center IPMN classification utilizing a comprehensive pancreas MRI dataset. This dataset includes 653 T1-weighted and 656 T2-weighted MRI images, accompanied by corresponding IPMN risk scores from 7 leading medical institutions, making it the largest and most diverse dataset for IPMN classification to date. We assess the performance of DenseNet-121 in both centralized and federated settings for training on distributed data. Our results demonstrate that the federated learning approach achieves high classification accuracy comparable to centralized learning while ensuring data privacy across institutions. This work marks a significant advancement in collaborative IPMN classification, facilitating secure and high-accuracy model training across multiple centers.
Adaptive Aggregation Weights for Federated Segmentation of Pancreas MRI
Oct 29, 2024

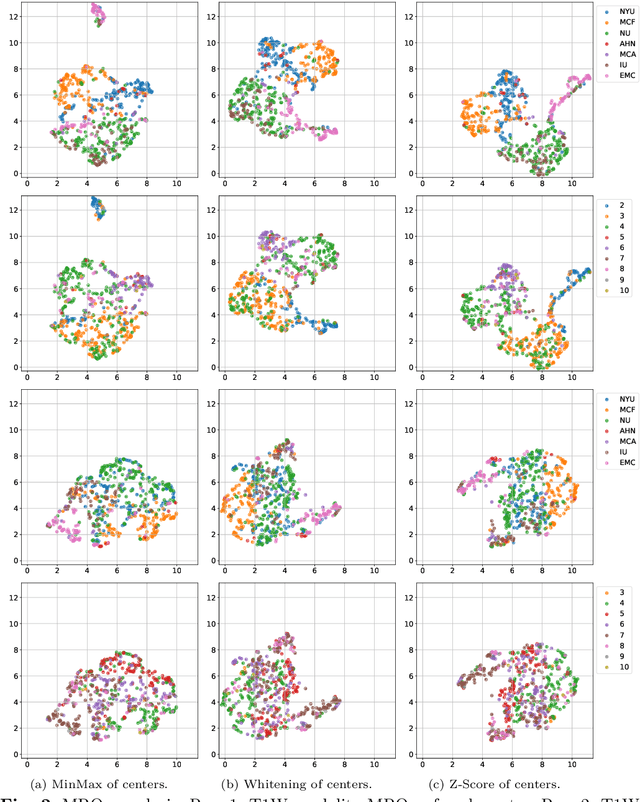
Abstract:Federated learning (FL) enables collaborative model training across institutions without sharing sensitive data, making it an attractive solution for medical imaging tasks. However, traditional FL methods, such as Federated Averaging (FedAvg), face difficulties in generalizing across domains due to variations in imaging protocols and patient demographics across institutions. This challenge is particularly evident in pancreas MRI segmentation, where anatomical variability and imaging artifacts significantly impact performance. In this paper, we conduct a comprehensive evaluation of FL algorithms for pancreas MRI segmentation and introduce a novel approach that incorporates adaptive aggregation weights. By dynamically adjusting the contribution of each client during model aggregation, our method accounts for domain-specific differences and improves generalization across heterogeneous datasets. Experimental results demonstrate that our approach enhances segmentation accuracy and reduces the impact of domain shift compared to conventional FL methods while maintaining privacy-preserving capabilities. Significant performance improvements are observed across multiple hospitals (centers).
Large-Scale Multi-Center CT and MRI Segmentation of Pancreas with Deep Learning
May 20, 2024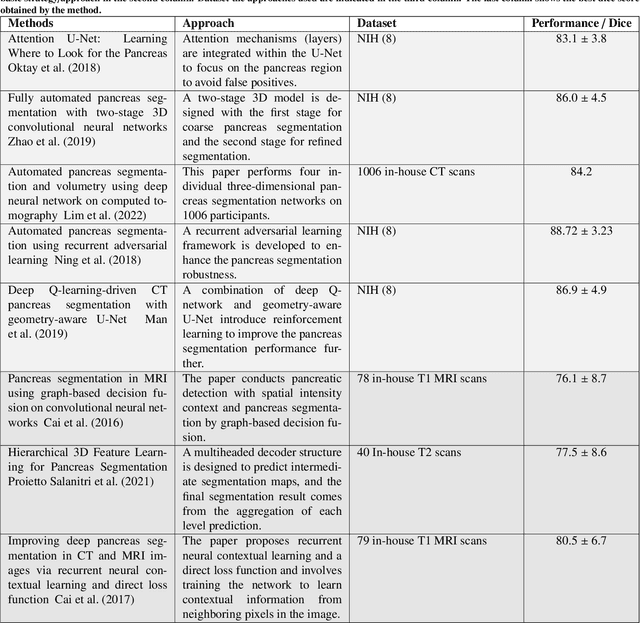
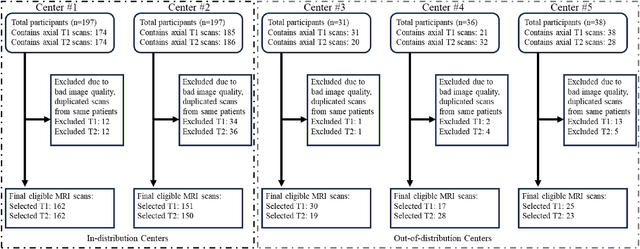
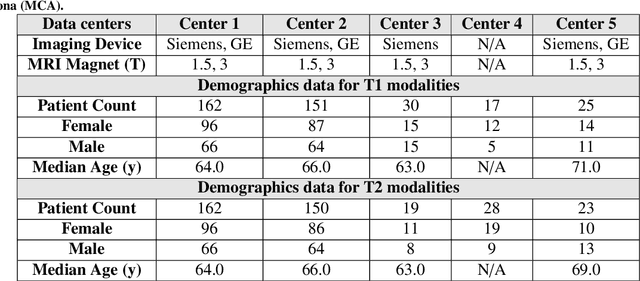
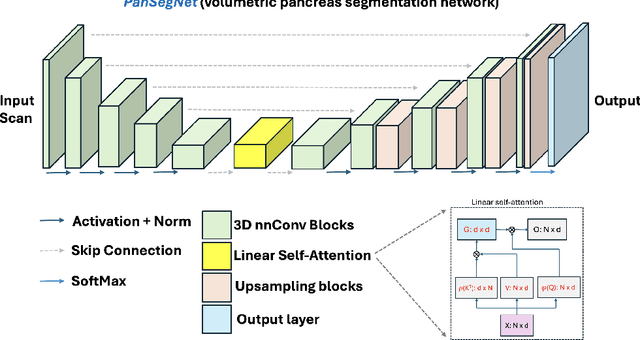
Abstract:Automated volumetric segmentation of the pancreas on cross-sectional imaging is needed for diagnosis and follow-up of pancreatic diseases. While CT-based pancreatic segmentation is more established, MRI-based segmentation methods are understudied, largely due to a lack of publicly available datasets, benchmarking research efforts, and domain-specific deep learning methods. In this retrospective study, we collected a large dataset (767 scans from 499 participants) of T1-weighted (T1W) and T2-weighted (T2W) abdominal MRI series from five centers between March 2004 and November 2022. We also collected CT scans of 1,350 patients from publicly available sources for benchmarking purposes. We developed a new pancreas segmentation method, called PanSegNet, combining the strengths of nnUNet and a Transformer network with a new linear attention module enabling volumetric computation. We tested PanSegNet's accuracy in cross-modality (a total of 2,117 scans) and cross-center settings with Dice and Hausdorff distance (HD95) evaluation metrics. We used Cohen's kappa statistics for intra and inter-rater agreement evaluation and paired t-tests for volume and Dice comparisons, respectively. For segmentation accuracy, we achieved Dice coefficients of 88.3% (std: 7.2%, at case level) with CT, 85.0% (std: 7.9%) with T1W MRI, and 86.3% (std: 6.4%) with T2W MRI. There was a high correlation for pancreas volume prediction with R^2 of 0.91, 0.84, and 0.85 for CT, T1W, and T2W, respectively. We found moderate inter-observer (0.624 and 0.638 for T1W and T2W MRI, respectively) and high intra-observer agreement scores. All MRI data is made available at https://osf.io/kysnj/. Our source code is available at https://github.com/NUBagciLab/PaNSegNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge