Gorkem Durak
CTest-Metric: A Unified Framework to Assess Clinical Validity of Metrics for CT Report Generation
Jan 16, 2026Abstract:In the generative AI era, where even critical medical tasks are increasingly automated, radiology report generation (RRG) continues to rely on suboptimal metrics for quality assessment. Developing domain-specific metrics has therefore been an active area of research, yet it remains challenging due to the lack of a unified, well-defined framework to assess their robustness and applicability in clinical contexts. To address this, we present CTest-Metric, a first unified metric assessment framework with three modules determining the clinical feasibility of metrics for CT RRG. The modules test: (i) Writing Style Generalizability (WSG) via LLM-based rephrasing; (ii) Synthetic Error Injection (SEI) at graded severities; and (iii) Metrics-vs-Expert correlation (MvE) using clinician ratings on 175 "disagreement" cases. Eight widely used metrics (BLEU, ROUGE, METEOR, BERTScore-F1, F1-RadGraph, RaTEScore, GREEN Score, CRG) are studied across seven LLMs built on a CT-CLIP encoder. Using our novel framework, we found that lexical NLG metrics are highly sensitive to stylistic variations; GREEN Score aligns best with expert judgments (Spearman~0.70), while CRG shows negative correlation; and BERTScore-F1 is least sensitive to factual error injection. We will release the framework, code, and allowable portion of the anonymized evaluation data (rephrased/error-injected CT reports), to facilitate reproducible benchmarking and future metric development.
ProDM: Synthetic Reality-driven Property-aware Progressive Diffusion Model for Coronary Calcium Motion Correction in Non-gated Chest CT
Dec 31, 2025Abstract:Coronary artery calcium (CAC) scoring from chest CT is a well-established tool to stratify and refine clinical cardiovascular disease risk estimation. CAC quantification relies on the accurate delineation of calcified lesions, but is oftentimes affected by artifacts introduced by cardiac and respiratory motion. ECG-gated cardiac CTs substantially reduce motion artifacts, but their use in population screening and routine imaging remains limited due to gating requirements and lack of insurance coverage. Although identification of incidental CAC from non-gated chest CT is increasingly considered for it offers an accessible and widely available alternative, this modality is limited by more severe motion artifacts. We present ProDM (Property-aware Progressive Correction Diffusion Model), a generative diffusion framework that restores motion-free calcified lesions from non-gated CTs. ProDM introduces three key components: (1) a CAC motion simulation data engine that synthesizes realistic non-gated acquisitions with diverse motion trajectories directly from cardiac-gated CTs, enabling supervised training without paired data; (2) a property-aware learning strategy incorporating calcium-specific priors through a differentiable calcium consistency loss to preserve lesion integrity; and (3) a progressive correction scheme that reduces artifacts gradually across diffusion steps to enhance stability and calcium fidelity. Experiments on real patient datasets show that ProDM significantly improves CAC scoring accuracy, spatial lesion fidelity, and risk stratification performance compared with several baselines. A reader study on real non-gated scans further confirms that ProDM suppresses motion artifacts and improves clinical usability. These findings highlight the potential of progressive, property-aware frameworks for reliable CAC quantification from routine chest CT imaging.
REN: Anatomically-Informed Mixture-of-Experts for Interstitial Lung Disease Diagnosis
Oct 06, 2025Abstract:Mixture-of-Experts (MoE) architectures have significantly contributed to scalable machine learning by enabling specialized subnetworks to tackle complex tasks efficiently. However, traditional MoE systems lack domain-specific constraints essential for medical imaging, where anatomical structure and regional disease heterogeneity strongly influence pathological patterns. Here, we introduce Regional Expert Networks (REN), the first anatomically-informed MoE framework tailored specifically for medical image classification. REN leverages anatomical priors to train seven specialized experts, each dedicated to distinct lung lobes and bilateral lung combinations, enabling precise modeling of region-specific pathological variations. Multi-modal gating mechanisms dynamically integrate radiomics biomarkers and deep learning (DL) features (CNN, ViT, Mamba) to weight expert contributions optimally. Applied to interstitial lung disease (ILD) classification, REN achieves consistently superior performance: the radiomics-guided ensemble reached an average AUC of 0.8646 +/- 0.0467, a +12.5 percent improvement over the SwinUNETR baseline (AUC 0.7685, p = 0.031). Region-specific experts further revealed that lower-lobe models achieved AUCs of 0.88-0.90, surpassing DL counterparts (CNN: 0.76-0.79) and aligning with known disease progression patterns. Through rigorous patient-level cross-validation, REN demonstrates strong generalizability and clinical interpretability, presenting a scalable, anatomically-guided approach readily extensible to other structured medical imaging applications.
SRMA-Mamba: Spatial Reverse Mamba Attention Network for Pathological Liver Segmentation in MRI Volumes
Aug 17, 2025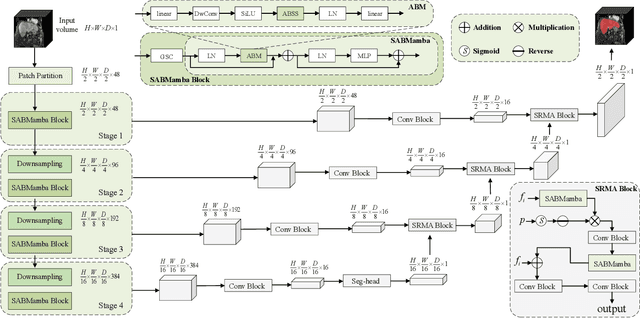
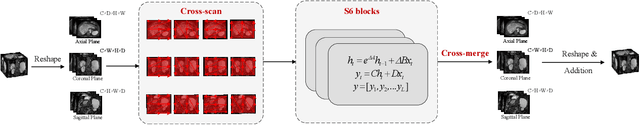
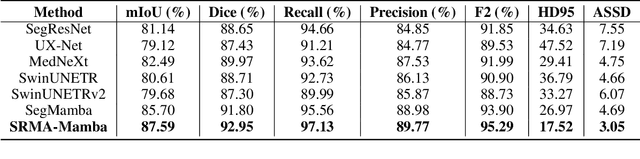
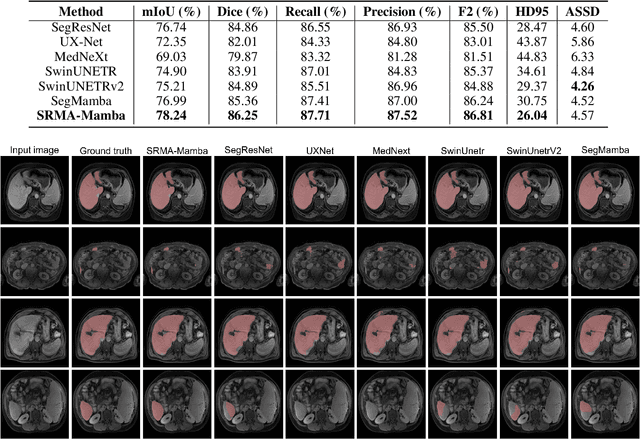
Abstract:Liver Cirrhosis plays a critical role in the prognosis of chronic liver disease. Early detection and timely intervention are critical in significantly reducing mortality rates. However, the intricate anatomical architecture and diverse pathological changes of liver tissue complicate the accurate detection and characterization of lesions in clinical settings. Existing methods underutilize the spatial anatomical details in volumetric MRI data, thereby hindering their clinical effectiveness and explainability. To address this challenge, we introduce a novel Mamba-based network, SRMA-Mamba, designed to model the spatial relationships within the complex anatomical structures of MRI volumes. By integrating the Spatial Anatomy-Based Mamba module (SABMamba), SRMA-Mamba performs selective Mamba scans within liver cirrhotic tissues and combines anatomical information from the sagittal, coronal, and axial planes to construct a global spatial context representation, enabling efficient volumetric segmentation of pathological liver structures. Furthermore, we introduce the Spatial Reverse Attention module (SRMA), designed to progressively refine cirrhotic details in the segmentation map, utilizing both the coarse segmentation map and hierarchical encoding features. Extensive experiments demonstrate that SRMA-Mamba surpasses state-of-the-art methods, delivering exceptional performance in 3D pathological liver segmentation. Our code is available for public: {\color{blue}{https://github.com/JunZengz/SRMA-Mamba}}.
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025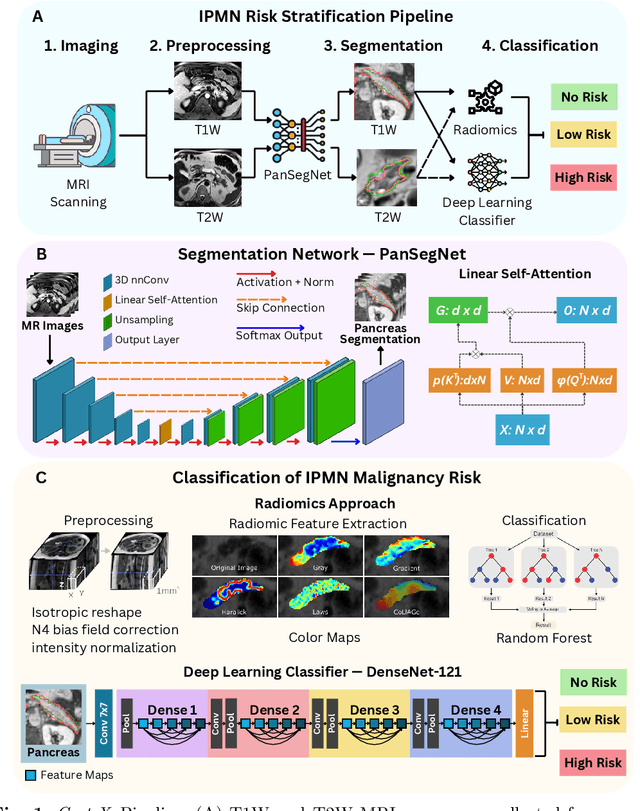

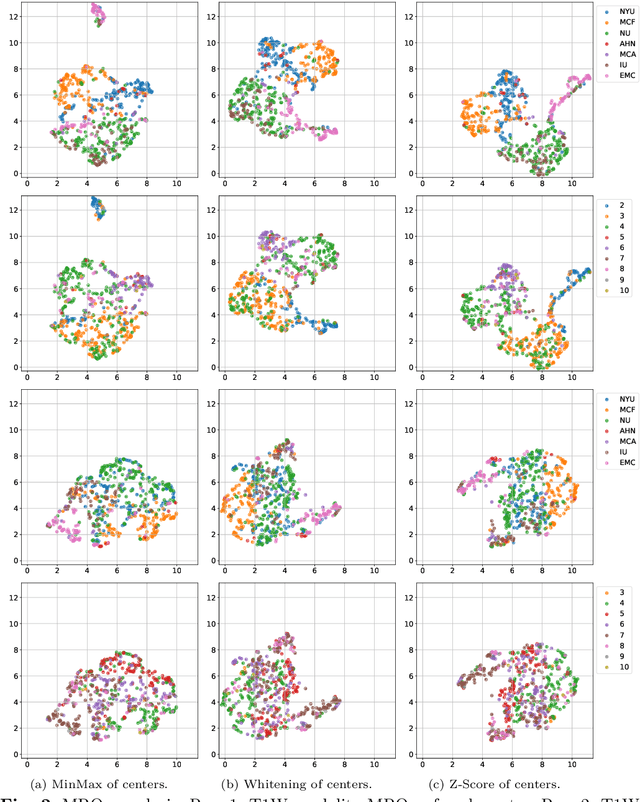
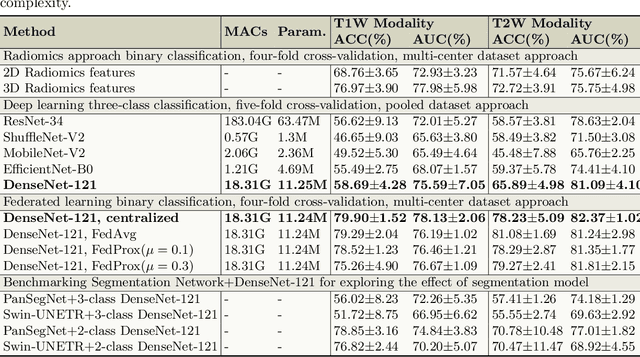
Abstract:Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
TAGS: 3D Tumor-Adaptive Guidance for SAM
May 21, 2025Abstract:Foundation models (FMs) such as CLIP and SAM have recently shown great promise in image segmentation tasks, yet their adaptation to 3D medical imaging-particularly for pathology detection and segmentation-remains underexplored. A critical challenge arises from the domain gap between natural images and medical volumes: existing FMs, pre-trained on 2D data, struggle to capture 3D anatomical context, limiting their utility in clinical applications like tumor segmentation. To address this, we propose an adaptation framework called TAGS: Tumor Adaptive Guidance for SAM, which unlocks 2D FMs for 3D medical tasks through multi-prompt fusion. By preserving most of the pre-trained weights, our approach enhances SAM's spatial feature extraction using CLIP's semantic insights and anatomy-specific prompts. Extensive experiments on three open-source tumor segmentation datasets prove that our model surpasses the state-of-the-art medical image segmentation models (+46.88% over nnUNet), interactive segmentation frameworks, and other established medical FMs, including SAM-Med2D, SAM-Med3D, SegVol, Universal, 3D-Adapter, and SAM-B (at least +13% over them). This highlights the robustness and adaptability of our proposed framework across diverse medical segmentation tasks.
Predicting Risk of Pulmonary Fibrosis Formation in PASC Patients
May 15, 2025Abstract:While the acute phase of the COVID-19 pandemic has subsided, its long-term effects persist through Post-Acute Sequelae of COVID-19 (PASC), commonly known as Long COVID. There remains substantial uncertainty regarding both its duration and optimal management strategies. PASC manifests as a diverse array of persistent or newly emerging symptoms--ranging from fatigue, dyspnea, and neurologic impairments (e.g., brain fog), to cardiovascular, pulmonary, and musculoskeletal abnormalities--that extend beyond the acute infection phase. This heterogeneous presentation poses substantial challenges for clinical assessment, diagnosis, and treatment planning. In this paper, we focus on imaging findings that may suggest fibrotic damage in the lungs, a critical manifestation characterized by scarring of lung tissue, which can potentially affect long-term respiratory function in patients with PASC. This study introduces a novel multi-center chest CT analysis framework that combines deep learning and radiomics for fibrosis prediction. Our approach leverages convolutional neural networks (CNNs) and interpretable feature extraction, achieving 82.2% accuracy and 85.5% AUC in classification tasks. We demonstrate the effectiveness of Grad-CAM visualization and radiomics-based feature analysis in providing clinically relevant insights for PASC-related lung fibrosis prediction. Our findings highlight the potential of deep learning-driven computational methods for early detection and risk assessment of PASC-related lung fibrosis--presented for the first time in the literature.
Shifts in Doctors' Eye Movements Between Real and AI-Generated Medical Images
Apr 21, 2025Abstract:Eye-tracking analysis plays a vital role in medical imaging, providing key insights into how radiologists visually interpret and diagnose clinical cases. In this work, we first analyze radiologists' attention and agreement by measuring the distribution of various eye-movement patterns, including saccades direction, amplitude, and their joint distribution. These metrics help uncover patterns in attention allocation and diagnostic strategies. Furthermore, we investigate whether and how doctors' gaze behavior shifts when viewing authentic (Real) versus deep-learning-generated (Fake) images. To achieve this, we examine fixation bias maps, focusing on first, last, short, and longest fixations independently, along with detailed saccades patterns, to quantify differences in gaze distribution and visual saliency between authentic and synthetic images.
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
Liver Cirrhosis Stage Estimation from MRI with Deep Learning
Feb 23, 2025Abstract:We present an end-to-end deep learning framework for automated liver cirrhosis stage estimation from multi-sequence MRI. Cirrhosis is the severe scarring (fibrosis) of the liver and a common endpoint of various chronic liver diseases. Early diagnosis is vital to prevent complications such as decompensation and cancer, which significantly decreases life expectancy. However, diagnosing cirrhosis in its early stages is challenging, and patients often present with life-threatening complications. Our approach integrates multi-scale feature learning with sequence-specific attention mechanisms to capture subtle tissue variations across cirrhosis progression stages. Using CirrMRI600+, a large-scale publicly available dataset of 628 high-resolution MRI scans from 339 patients, we demonstrate state-of-the-art performance in three-stage cirrhosis classification. Our best model achieves 72.8% accuracy on T1W and 63.8% on T2W sequences, significantly outperforming traditional radiomics-based approaches. Through extensive ablation studies, we show that our architecture effectively learns stage-specific imaging biomarkers. We establish new benchmarks for automated cirrhosis staging and provide insights for developing clinically applicable deep learning systems. The source code will be available at https://github.com/JunZengz/CirrhosisStage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge