Abhishek Srivastava
Is Long Range Sequential Modeling Necessary For Colorectal Tumor Segmentation?
Feb 10, 2025Abstract:Segmentation of colorectal cancer (CRC) tumors in 3D medical imaging is both complex and clinically critical, providing vital support for effective radiation therapy planning and survival outcome assessment. Recently, 3D volumetric segmentation architectures incorporating long-range sequence modeling mechanisms, such as Transformers and Mamba, have gained attention for their capacity to achieve high accuracy in 3D medical image segmentation. In this work, we evaluate the effectiveness of these global token modeling techniques by pitting them against our proposed MambaOutUNet within the context of our newly introduced colorectal tumor segmentation dataset (CTS-204). Our findings suggest that robust local token interactions can outperform long-range modeling techniques in cases where the region of interest is small and anatomically complex, proposing a potential shift in 3D tumor segmentation research.
EB-NeRD: A Large-Scale Dataset for News Recommendation
Oct 04, 2024
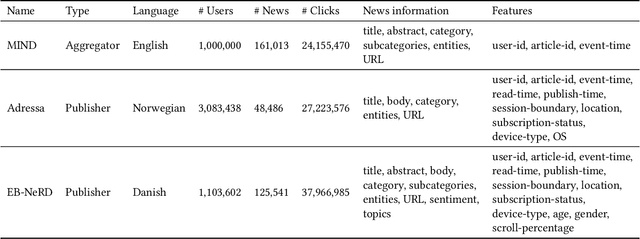

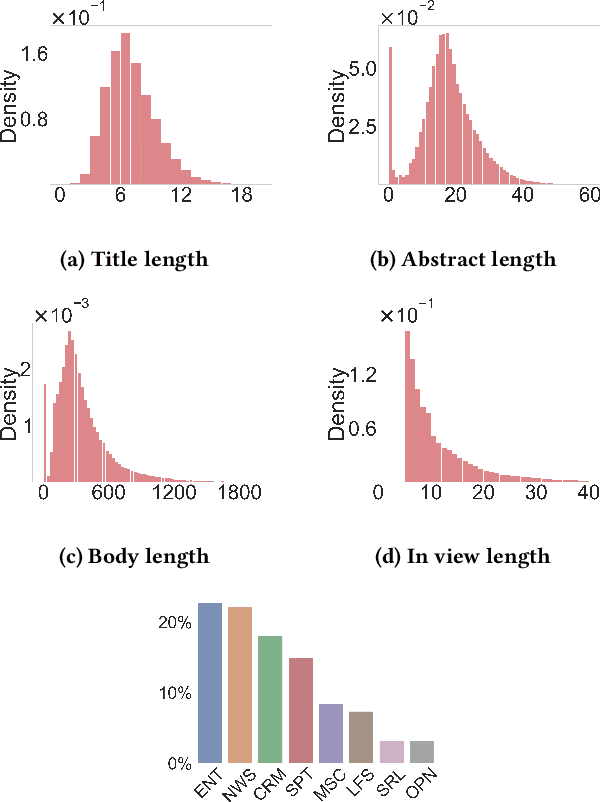
Abstract:Personalized content recommendations have been pivotal to the content experience in digital media from video streaming to social networks. However, several domain specific challenges have held back adoption of recommender systems in news publishing. To address these challenges, we introduce the Ekstra Bladet News Recommendation Dataset (EB-NeRD). The dataset encompasses data from over a million unique users and more than 37 million impression logs from Ekstra Bladet. It also includes a collection of over 125,000 Danish news articles, complete with titles, abstracts, bodies, and metadata, such as categories. EB-NeRD served as the benchmark dataset for the RecSys '24 Challenge, where it was demonstrated how the dataset can be used to address both technical and normative challenges in designing effective and responsible recommender systems for news publishing. The dataset is available at: https://recsys.eb.dk.
RecSys Challenge 2024: Balancing Accuracy and Editorial Values in News Recommendations
Sep 30, 2024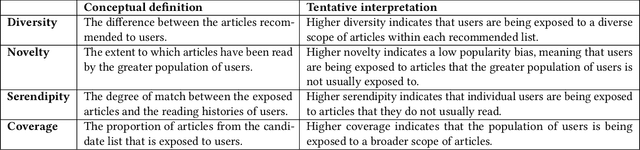
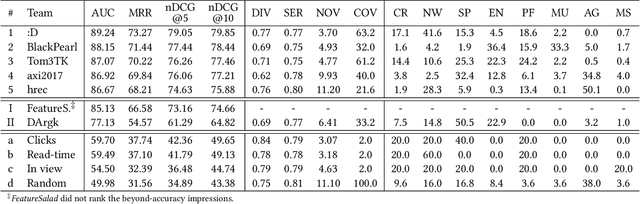
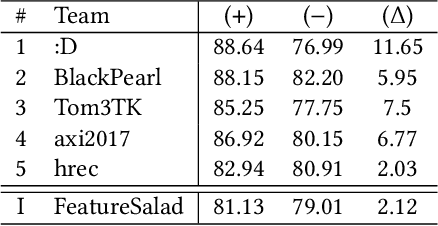
Abstract:The RecSys Challenge 2024 aims to advance news recommendation by addressing both the technical and normative challenges inherent in designing effective and responsible recommender systems for news publishing. This paper describes the challenge, including its objectives, problem setting, and the dataset provided by the Danish news publishers Ekstra Bladet and JP/Politikens Media Group ("Ekstra Bladet"). The challenge explores the unique aspects of news recommendation, such as modeling user preferences based on behavior, accounting for the influence of the news agenda on user interests, and managing the rapid decay of news items. Additionally, the challenge embraces normative complexities, investigating the effects of recommender systems on news flow and their alignment with editorial values. We summarize the challenge setup, dataset characteristics, and evaluation metrics. Finally, we announce the winners and highlight their contributions. The dataset is available at: https://recsys.eb.dk.
Multi-Scale Fusion Methodologies for Head and Neck Tumor Segmentation
Oct 29, 2022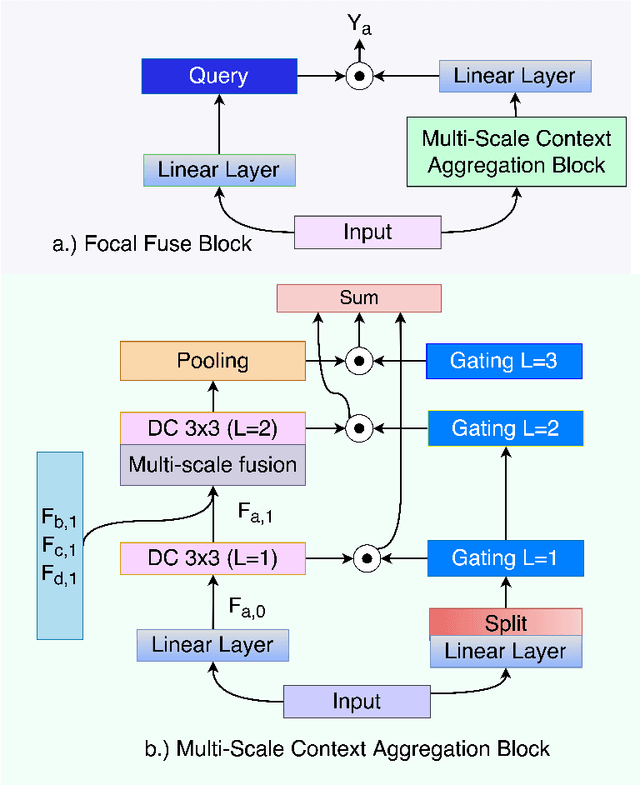

Abstract:Head and Neck (H\&N) organ-at-risk (OAR) and tumor segmentations are essential components of radiation therapy planning. The varying anatomic locations and dimensions of H\&N nodal Gross Tumor Volumes (GTVn) and H\&N primary gross tumor volume (GTVp) are difficult to obtain due to lack of accurate and reliable delineation methods. The downstream effect of incorrect segmentation can result in unnecessary irradiation of normal organs. Towards a fully automated radiation therapy planning algorithm, we explore the efficacy of multi-scale fusion based deep learning architectures for accurately segmenting H\&N tumors from medical scans.
An Efficient Multi-Scale Fusion Network for 3D Organ at Risk Segmentation
Aug 15, 2022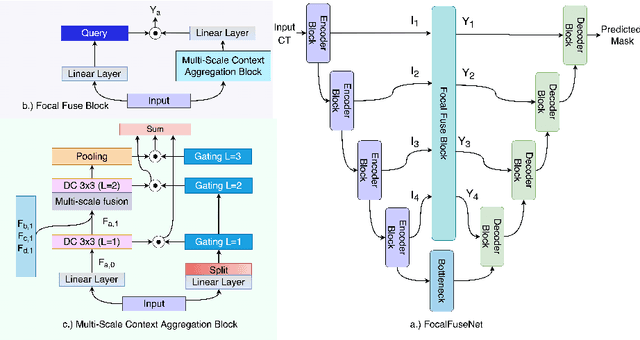
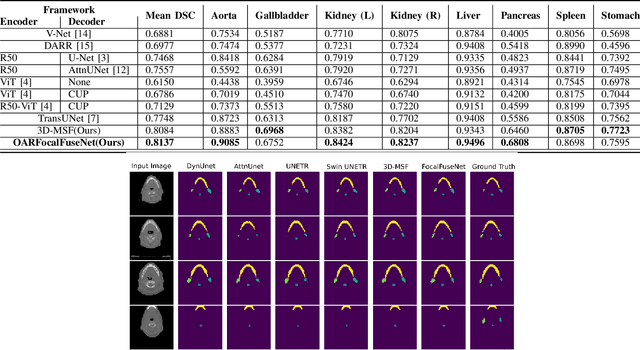
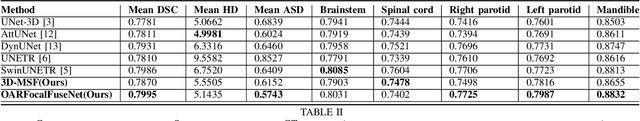
Abstract:Accurate segmentation of organs-at-risks (OARs) is a precursor for optimizing radiation therapy planning. Existing deep learning-based multi-scale fusion architectures have demonstrated a tremendous capacity for 2D medical image segmentation. The key to their success is aggregating global context and maintaining high resolution representations. However, when translated into 3D segmentation problems, existing multi-scale fusion architectures might underperform due to their heavy computation overhead and substantial data diet. To address this issue, we propose a new OAR segmentation framework, called OARFocalFuseNet, which fuses multi-scale features and employs focal modulation for capturing global-local context across multiple scales. Each resolution stream is enriched with features from different resolution scales, and multi-scale information is aggregated to model diverse contextual ranges. As a result, feature representations are further boosted. The comprehensive comparisons in our experimental setup with OAR segmentation as well as multi-organ segmentation show that our proposed OARFocalFuseNet outperforms the recent state-of-the-art methods on publicly available OpenKBP datasets and Synapse multi-organ segmentation. Both of the proposed methods (3D-MSF and OARFocalFuseNet) showed promising performance in terms of standard evaluation metrics. Our best performing method (OARFocalFuseNet) obtained a dice coefficient of 0.7995 and hausdorff distance of 5.1435 on OpenKBP datasets and dice coefficient of 0.8137 on Synapse multi-organ segmentation dataset.
Automatic Polyp Segmentation with Multiple Kernel Dilated Convolution Network
Jun 23, 2022



Abstract:The detection and removal of precancerous polyps through colonoscopy is the primary technique for the prevention of colorectal cancer worldwide. However, the miss rate of colorectal polyp varies significantly among the endoscopists. It is well known that a computer-aided diagnosis (CAD) system can assist endoscopists in detecting colon polyps and minimize the variation among endoscopists. In this study, we introduce a novel deep learning architecture, named MKDCNet, for automatic polyp segmentation robust to significant changes in polyp data distribution. MKDCNet is simply an encoder-decoder neural network that uses the pre-trained ResNet50 as the encoder and novel multiple kernel dilated convolution (MKDC) block that expands the field of view to learn more robust and heterogeneous representation. Extensive experiments on four publicly available polyp datasets and cell nuclei dataset show that the proposed MKDCNet outperforms the state-of-the-art methods when trained and tested on the same dataset as well when tested on unseen polyp datasets from different distributions. With rich results, we demonstrated the robustness of the proposed architecture. From an efficiency perspective, our algorithm can process at (approx 45) frames per second on RTX 3090 GPU. MKDCNet can be a strong benchmark for building real-time systems for clinical colonoscopies. The code of the proposed MKDCNet is available at https://github.com/nikhilroxtomar/MKDCNet.
Video Capsule Endoscopy Classification using Focal Modulation Guided Convolutional Neural Network
Jun 16, 2022



Abstract:Video capsule endoscopy is a hot topic in computer vision and medicine. Deep learning can have a positive impact on the future of video capsule endoscopy technology. It can improve the anomaly detection rate, reduce physicians' time for screening, and aid in real-world clinical analysis. CADx classification system for video capsule endoscopy has shown a great promise for further improvement. For example, detection of cancerous polyp and bleeding can lead to swift medical response and improve the survival rate of the patients. To this end, an automated CADx system must have high throughput and decent accuracy. In this paper, we propose FocalConvNet, a focal modulation network integrated with lightweight convolutional layers for the classification of small bowel anatomical landmarks and luminal findings. FocalConvNet leverages focal modulation to attain global context and allows global-local spatial interactions throughout the forward pass. Moreover, the convolutional block with its intrinsic inductive/learning bias and capacity to extract hierarchical features allows our FocalConvNet to achieve favourable results with high throughput. We compare our FocalConvNet with other SOTA on Kvasir-Capsule, a large-scale VCE dataset with 44,228 frames with 13 classes of different anomalies. Our proposed method achieves the weighted F1-score, recall and MCC} of 0.6734, 0.6373 and 0.2974, respectively outperforming other SOTA methodologies. Furthermore, we report the highest throughput of 148.02 images/second rate to establish the potential of FocalConvNet in a real-time clinical environment. The code of the proposed FocalConvNet is available at https://github.com/NoviceMAn-prog/FocalConvNet.
Challenges and Solutions to Build a Data Pipeline to Identify Anomalies in Enterprise System Performance
Dec 13, 2021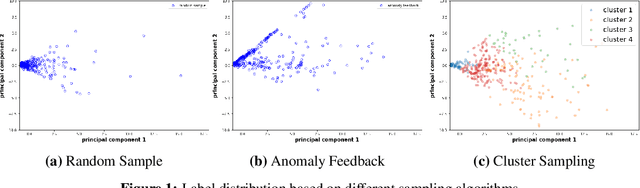
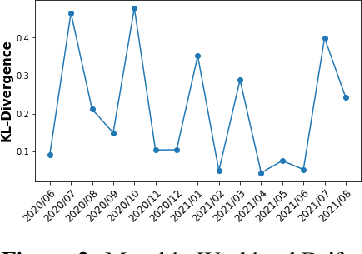
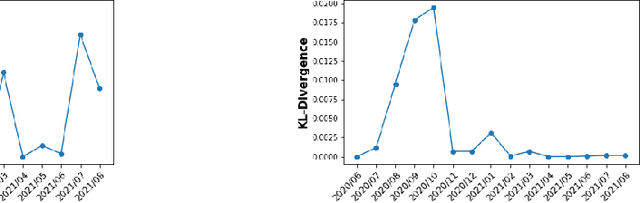
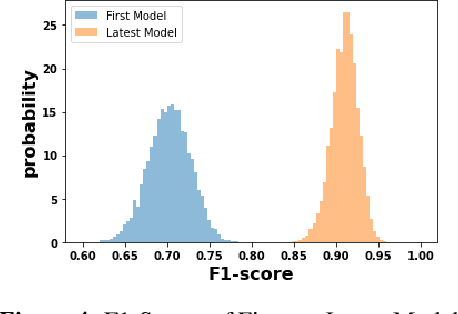
Abstract:We discuss how VMware is solving the following challenges to harness data to operate our ML-based anomaly detection system to detect performance issues in our Software Defined Data Center (SDDC) enterprise deployments: (i) label scarcity and label bias due to heavy dependency on unscalable human annotators, and (ii) data drifts due to ever-changing workload patterns, software stack and underlying hardware. Our anomaly detection system has been deployed in production for many years and has successfully detected numerous major performance issues. We demonstrate that by addressing these data challenges, we not only improve the accuracy of our performance anomaly detection model by 30%, but also ensure that the model performance to never degrade over time.
PAANet: Progressive Alternating Attention for Automatic Medical Image Segmentation
Nov 20, 2021



Abstract:Medical image segmentation can provide detailed information for clinical analysis which can be useful for scenarios where the detailed location of a finding is important. Knowing the location of disease can play a vital role in treatment and decision-making. Convolutional neural network (CNN) based encoder-decoder techniques have advanced the performance of automated medical image segmentation systems. Several such CNN-based methodologies utilize techniques such as spatial- and channel-wise attention to enhance performance. Another technique that has drawn attention in recent years is residual dense blocks (RDBs). The successive convolutional layers in densely connected blocks are capable of extracting diverse features with varied receptive fields and thus, enhancing performance. However, consecutive stacked convolutional operators may not necessarily generate features that facilitate the identification of the target structures. In this paper, we propose a progressive alternating attention network (PAANet). We develop progressive alternating attention dense (PAAD) blocks, which construct a guiding attention map (GAM) after every convolutional layer in the dense blocks using features from all scales. The GAM allows the following layers in the dense blocks to focus on the spatial locations relevant to the target region. Every alternate PAAD block inverts the GAM to generate a reverse attention map which guides ensuing layers to extract boundary and edge-related information, refining the segmentation process. Our experiments on three different biomedical image segmentation datasets exhibit that our PAANet achieves favourable performance when compared to other state-of-the-art methods.
GMSRF-Net: An improved generalizability with global multi-scale residual fusion network for polyp segmentation
Nov 20, 2021



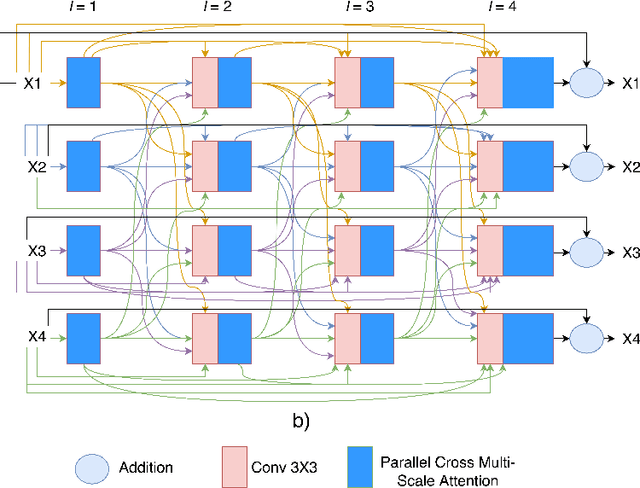
Abstract:Colonoscopy is a gold standard procedure but is highly operator-dependent. Efforts have been made to automate the detection and segmentation of polyps, a precancerous precursor, to effectively minimize missed rate. Widely used computer-aided polyp segmentation systems actuated by encoder-decoder have achieved high performance in terms of accuracy. However, polyp segmentation datasets collected from varied centers can follow different imaging protocols leading to difference in data distribution. As a result, most methods suffer from performance drop and require re-training for each specific dataset. We address this generalizability issue by proposing a global multi-scale residual fusion network (GMSRF-Net). Our proposed network maintains high-resolution representations while performing multi-scale fusion operations for all resolution scales. To further leverage scale information, we design cross multi-scale attention (CMSA) and multi-scale feature selection (MSFS) modules within the GMSRF-Net. The repeated fusion operations gated by CMSA and MSFS demonstrate improved generalizability of the network. Experiments conducted on two different polyp segmentation datasets show that our proposed GMSRF-Net outperforms the previous top-performing state-of-the-art method by 8.34% and 10.31% on unseen CVC-ClinicDB and unseen Kvasir-SEG, in terms of dice coefficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge