Mohamed Abazeed
Self-Supervised Learning for Organs At Risk and Tumor Segmentation with Uncertainty Quantification
May 04, 2023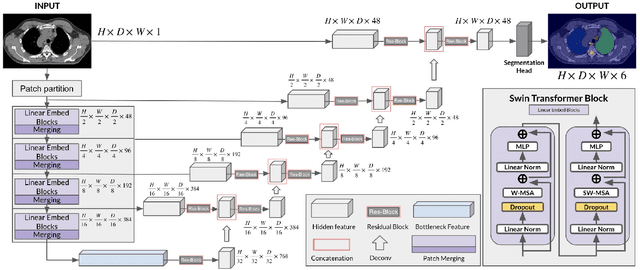
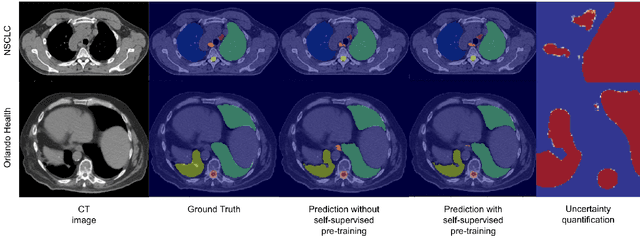
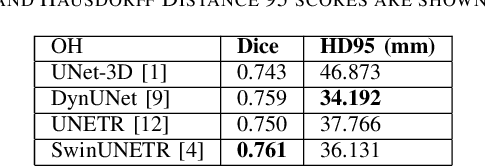
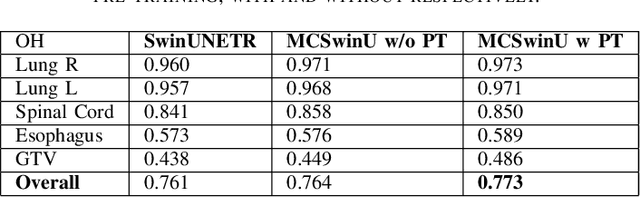
Abstract:In this study, our goal is to show the impact of self-supervised pre-training of transformers for organ at risk (OAR) and tumor segmentation as compared to costly fully-supervised learning. The proposed algorithm is called Monte Carlo Transformer based U-Net (MC-Swin-U). Unlike many other available models, our approach presents uncertainty quantification with Monte Carlo dropout strategy while generating its voxel-wise prediction. We test and validate the proposed model on both public and one private datasets and evaluate the gross tumor volume (GTV) as well as nearby risky organs' boundaries. We show that self-supervised pre-training approach improves the segmentation scores significantly while providing additional benefits for avoiding large-scale annotation costs.
An Efficient Multi-Scale Fusion Network for 3D Organ at Risk Segmentation
Aug 15, 2022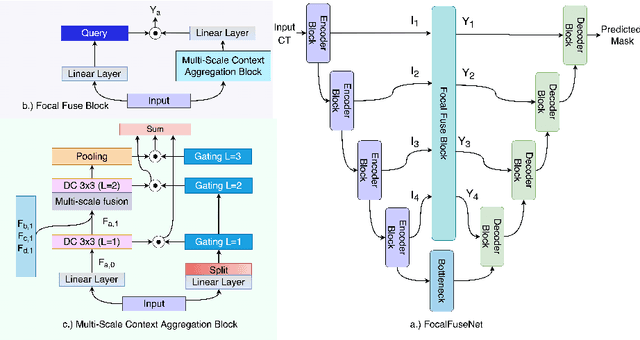
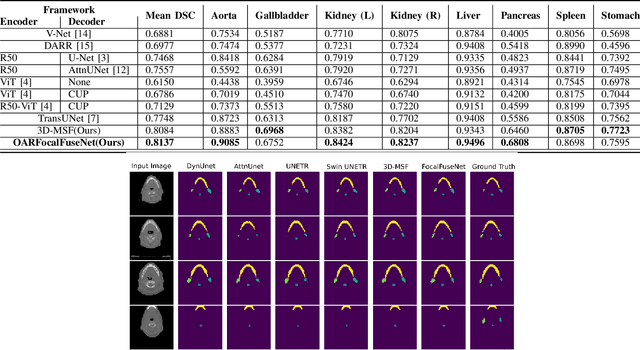
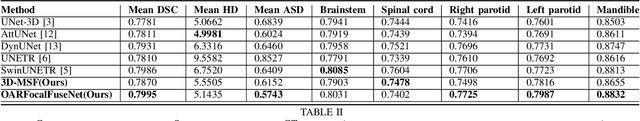
Abstract:Accurate segmentation of organs-at-risks (OARs) is a precursor for optimizing radiation therapy planning. Existing deep learning-based multi-scale fusion architectures have demonstrated a tremendous capacity for 2D medical image segmentation. The key to their success is aggregating global context and maintaining high resolution representations. However, when translated into 3D segmentation problems, existing multi-scale fusion architectures might underperform due to their heavy computation overhead and substantial data diet. To address this issue, we propose a new OAR segmentation framework, called OARFocalFuseNet, which fuses multi-scale features and employs focal modulation for capturing global-local context across multiple scales. Each resolution stream is enriched with features from different resolution scales, and multi-scale information is aggregated to model diverse contextual ranges. As a result, feature representations are further boosted. The comprehensive comparisons in our experimental setup with OAR segmentation as well as multi-organ segmentation show that our proposed OARFocalFuseNet outperforms the recent state-of-the-art methods on publicly available OpenKBP datasets and Synapse multi-organ segmentation. Both of the proposed methods (3D-MSF and OARFocalFuseNet) showed promising performance in terms of standard evaluation metrics. Our best performing method (OARFocalFuseNet) obtained a dice coefficient of 0.7995 and hausdorff distance of 5.1435 on OpenKBP datasets and dice coefficient of 0.8137 on Synapse multi-organ segmentation dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge