Elif Keles
Ensemble Models for Predicting Treatment Response in Pediatric Low-Grade Glioma Managed with Chemotherapy
Jan 07, 2026Abstract:In this paper, we introduce a novel pipeline for predicting chemotherapy response in pediatric brain tumors that are not amenable to complete surgical resection, using pre-treatment magnetic resonance imaging combined with clinical information. Our method integrates a state-of-the-art pediatric brain tumor segmentation framework with radiomic feature extraction and clinical data through an ensemble of a Swin UNETR encoder and XGBoost classifier. The segmentation model delineates four tumor subregions enhancing tumor, non-enhancing tumor, cystic component and edema which are used to extract imaging biomarkers and generate predictive features. The Swin UNETR network classifies the response to treatment directly from these segmented MRI scans, while XGBoost predicts response using radiomics and clinical variables including legal sex, ethnicity, race, age at event (in days), molecular subtype, tumor locations, initial surgery status, metastatic status, metastasis location, chemotherapy type, protocol name and chemotherapy agents. The ensemble output provides a non-invasive estimate of chemotherapy response in this historically challenging population characterized by lower progression-free survival. Among compared approaches, our Swin-Ensemble achieved the best performance (precision for non effective cases=0.68, recall for non effective cases=0.85, precision for chemotherapy effective cases=0.64 and overall accuracy=0.69), outperforming Mamba-FeatureFuse, Swin UNETR encoder, and Swin-FeatureFuse models. Our findings suggest that this ensemble framework represents a promising step toward personalized therapy response prediction for pediatric low-grade glioma patients in need of chemotherapy treatment who are not suitable for complete surgical resection, a population with significantly lower progression free survival and for whom chemotherapy remains the primary treatment option.
SRMA-Mamba: Spatial Reverse Mamba Attention Network for Pathological Liver Segmentation in MRI Volumes
Aug 17, 2025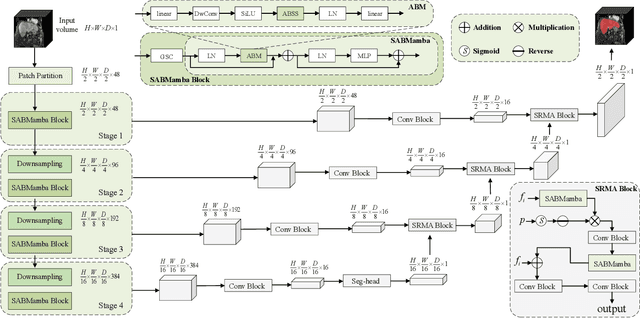
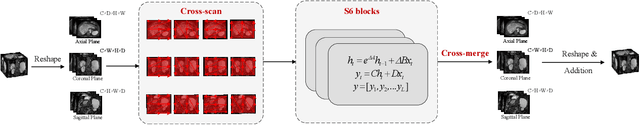
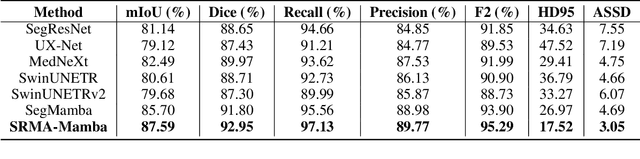
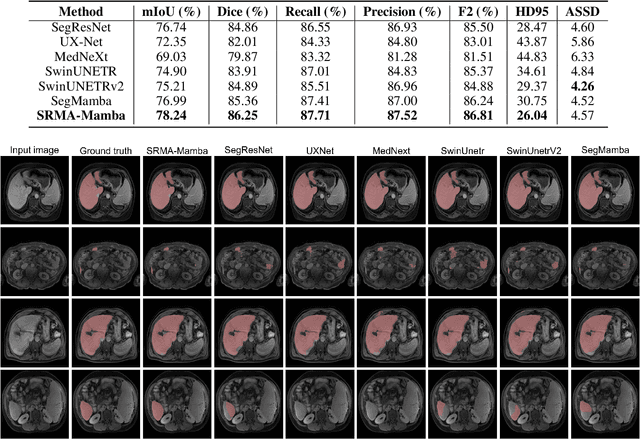
Abstract:Liver Cirrhosis plays a critical role in the prognosis of chronic liver disease. Early detection and timely intervention are critical in significantly reducing mortality rates. However, the intricate anatomical architecture and diverse pathological changes of liver tissue complicate the accurate detection and characterization of lesions in clinical settings. Existing methods underutilize the spatial anatomical details in volumetric MRI data, thereby hindering their clinical effectiveness and explainability. To address this challenge, we introduce a novel Mamba-based network, SRMA-Mamba, designed to model the spatial relationships within the complex anatomical structures of MRI volumes. By integrating the Spatial Anatomy-Based Mamba module (SABMamba), SRMA-Mamba performs selective Mamba scans within liver cirrhotic tissues and combines anatomical information from the sagittal, coronal, and axial planes to construct a global spatial context representation, enabling efficient volumetric segmentation of pathological liver structures. Furthermore, we introduce the Spatial Reverse Attention module (SRMA), designed to progressively refine cirrhotic details in the segmentation map, utilizing both the coarse segmentation map and hierarchical encoding features. Extensive experiments demonstrate that SRMA-Mamba surpasses state-of-the-art methods, delivering exceptional performance in 3D pathological liver segmentation. Our code is available for public: {\color{blue}{https://github.com/JunZengz/SRMA-Mamba}}.
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025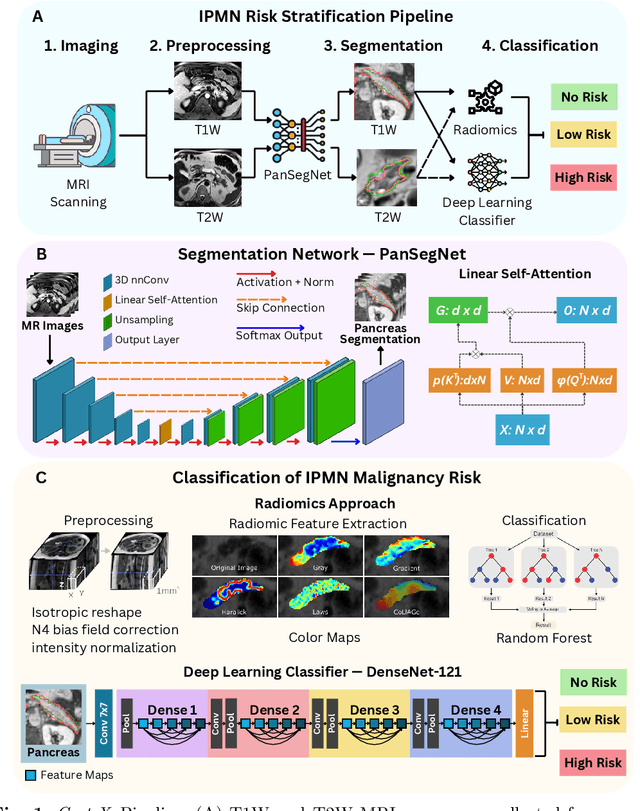

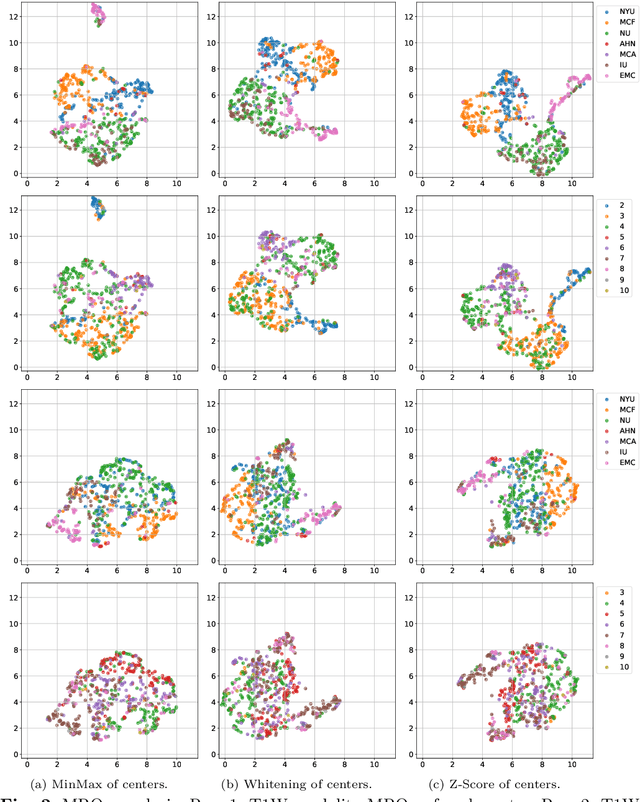
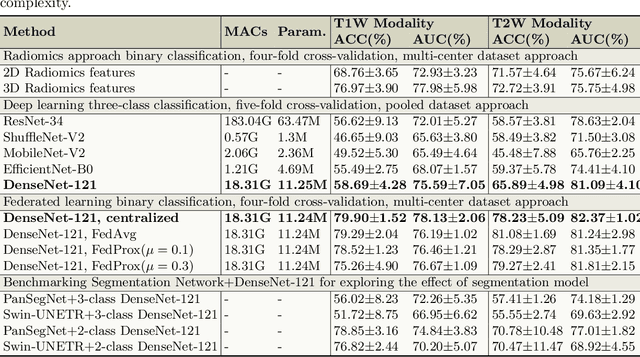
Abstract:Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
Predicting Risk of Pulmonary Fibrosis Formation in PASC Patients
May 15, 2025Abstract:While the acute phase of the COVID-19 pandemic has subsided, its long-term effects persist through Post-Acute Sequelae of COVID-19 (PASC), commonly known as Long COVID. There remains substantial uncertainty regarding both its duration and optimal management strategies. PASC manifests as a diverse array of persistent or newly emerging symptoms--ranging from fatigue, dyspnea, and neurologic impairments (e.g., brain fog), to cardiovascular, pulmonary, and musculoskeletal abnormalities--that extend beyond the acute infection phase. This heterogeneous presentation poses substantial challenges for clinical assessment, diagnosis, and treatment planning. In this paper, we focus on imaging findings that may suggest fibrotic damage in the lungs, a critical manifestation characterized by scarring of lung tissue, which can potentially affect long-term respiratory function in patients with PASC. This study introduces a novel multi-center chest CT analysis framework that combines deep learning and radiomics for fibrosis prediction. Our approach leverages convolutional neural networks (CNNs) and interpretable feature extraction, achieving 82.2% accuracy and 85.5% AUC in classification tasks. We demonstrate the effectiveness of Grad-CAM visualization and radiomics-based feature analysis in providing clinically relevant insights for PASC-related lung fibrosis prediction. Our findings highlight the potential of deep learning-driven computational methods for early detection and risk assessment of PASC-related lung fibrosis--presented for the first time in the literature.
Liver Cirrhosis Stage Estimation from MRI with Deep Learning
Feb 23, 2025Abstract:We present an end-to-end deep learning framework for automated liver cirrhosis stage estimation from multi-sequence MRI. Cirrhosis is the severe scarring (fibrosis) of the liver and a common endpoint of various chronic liver diseases. Early diagnosis is vital to prevent complications such as decompensation and cancer, which significantly decreases life expectancy. However, diagnosing cirrhosis in its early stages is challenging, and patients often present with life-threatening complications. Our approach integrates multi-scale feature learning with sequence-specific attention mechanisms to capture subtle tissue variations across cirrhosis progression stages. Using CirrMRI600+, a large-scale publicly available dataset of 628 high-resolution MRI scans from 339 patients, we demonstrate state-of-the-art performance in three-stage cirrhosis classification. Our best model achieves 72.8% accuracy on T1W and 63.8% on T2W sequences, significantly outperforming traditional radiomics-based approaches. Through extensive ablation studies, we show that our architecture effectively learns stage-specific imaging biomarkers. We establish new benchmarks for automated cirrhosis staging and provide insights for developing clinically applicable deep learning systems. The source code will be available at https://github.com/JunZengz/CirrhosisStage.
Mortality Prediction of Pulmonary Embolism Patients with Deep Learning and XGBoost
Nov 27, 2024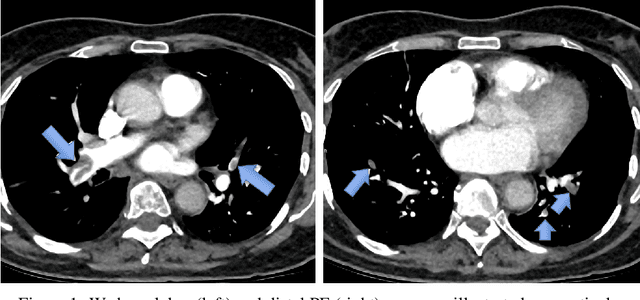

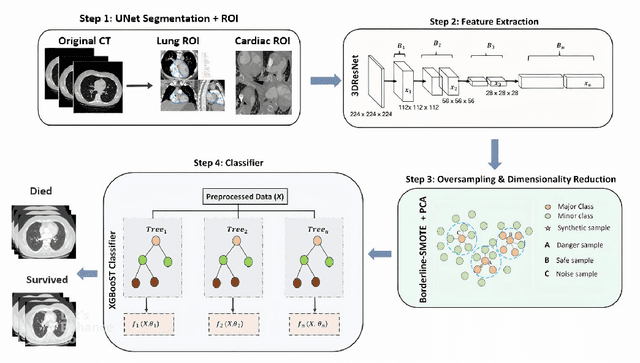
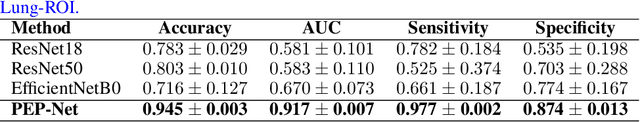
Abstract:Pulmonary Embolism (PE) is a serious cardiovascular condition that remains a leading cause of mortality and critical illness, underscoring the need for enhanced diagnostic strategies. Conventional clinical methods have limited success in predicting 30-day in-hospital mortality of PE patients. In this study, we present a new algorithm, called PEP-Net, for 30-day mortality prediction of PE patients based on the initial imaging data (CT) that opportunistically integrates a 3D Residual Network (3DResNet) with Extreme Gradient Boosting (XGBoost) algorithm with patient level binary labels without annotations of the emboli and its extent. Our proposed system offers a comprehensive prediction strategy by handling class imbalance problems, reducing overfitting via regularization, and reducing the prediction variance for more stable predictions. PEP-Net was tested in a cohort of 193 volumetric CT scans diagnosed with Acute PE, and it demonstrated a superior performance by significantly outperforming baseline models (76-78\%) with an accuracy of 94.5\% (+/-0.3) and 94.0\% (+/-0.7) when the input image is either lung region (Lung-ROI) or heart region (Cardiac-ROI). Our results advance PE prognostics by using only initial imaging data, setting a new benchmark in the field. While purely deep learning models have become the go-to for many medical classification (diagnostic) tasks, combined ResNet and XGBoost models herein outperform sole deep learning models due to a potential reason for having lack of enough data.
IPMN Risk Assessment under Federated Learning Paradigm
Nov 08, 2024



Abstract:Accurate classification of Intraductal Papillary Mucinous Neoplasms (IPMN) is essential for identifying high-risk cases that require timely intervention. In this study, we develop a federated learning framework for multi-center IPMN classification utilizing a comprehensive pancreas MRI dataset. This dataset includes 653 T1-weighted and 656 T2-weighted MRI images, accompanied by corresponding IPMN risk scores from 7 leading medical institutions, making it the largest and most diverse dataset for IPMN classification to date. We assess the performance of DenseNet-121 in both centralized and federated settings for training on distributed data. Our results demonstrate that the federated learning approach achieves high classification accuracy comparable to centralized learning while ensuring data privacy across institutions. This work marks a significant advancement in collaborative IPMN classification, facilitating secure and high-accuracy model training across multiple centers.
A New Logic For Pediatric Brain Tumor Segmentation
Nov 03, 2024



Abstract:In this paper, we present a novel approach for segmenting pediatric brain tumors using a deep learning architecture, inspired by expert radiologists' segmentation strategies. Our model delineates four distinct tumor labels and is benchmarked on a held-out PED BraTS 2024 test set (i.e., pediatric brain tumor datasets introduced by BraTS). Furthermore, we evaluate our model's performance against the state-of-the-art (SOTA) model using a new external dataset of 30 patients from CBTN (Children's Brain Tumor Network), labeled in accordance with the PED BraTS 2024 guidelines. We compare segmentation outcomes with the winning algorithm from the PED BraTS 2023 challenge as the SOTA model. Our proposed algorithm achieved an average Dice score of 0.642 and an HD95 of 73.0 mm on the CBTN test data, outperforming the SOTA model, which achieved a Dice score of 0.626 and an HD95 of 84.0 mm. Our results indicate that the proposed model is a step towards providing more accurate segmentation for pediatric brain tumors, which is essential for evaluating therapy response and monitoring patient progress.
Adaptive Aggregation Weights for Federated Segmentation of Pancreas MRI
Oct 29, 2024

Abstract:Federated learning (FL) enables collaborative model training across institutions without sharing sensitive data, making it an attractive solution for medical imaging tasks. However, traditional FL methods, such as Federated Averaging (FedAvg), face difficulties in generalizing across domains due to variations in imaging protocols and patient demographics across institutions. This challenge is particularly evident in pancreas MRI segmentation, where anatomical variability and imaging artifacts significantly impact performance. In this paper, we conduct a comprehensive evaluation of FL algorithms for pancreas MRI segmentation and introduce a novel approach that incorporates adaptive aggregation weights. By dynamically adjusting the contribution of each client during model aggregation, our method accounts for domain-specific differences and improves generalization across heterogeneous datasets. Experimental results demonstrate that our approach enhances segmentation accuracy and reduces the impact of domain shift compared to conventional FL methods while maintaining privacy-preserving capabilities. Significant performance improvements are observed across multiple hospitals (centers).
Towards Synergistic Deep Learning Models for Volumetric Cirrhotic Liver Segmentation in MRIs
Aug 08, 2024



Abstract:Liver cirrhosis, a leading cause of global mortality, requires precise segmentation of ROIs for effective disease monitoring and treatment planning. Existing segmentation models often fail to capture complex feature interactions and generalize across diverse datasets. To address these limitations, we propose a novel synergistic theory that leverages complementary latent spaces for enhanced feature interaction modeling. Our proposed architecture, nnSynergyNet3D integrates continuous and discrete latent spaces for 3D volumes and features auto-configured training. This approach captures both fine-grained and coarse features, enabling effective modeling of intricate feature interactions. We empirically validated nnSynergyNet3D on a private dataset of 628 high-resolution T1 abdominal MRI scans from 339 patients. Our model outperformed the baseline nnUNet3D by approximately 2%. Additionally, zero-shot testing on healthy liver CT scans from the public LiTS dataset demonstrated superior cross-modal generalization capabilities. These results highlight the potential of synergistic latent space models to improve segmentation accuracy and robustness, thereby enhancing clinical workflows by ensuring consistency across CT and MRI modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge