Marius G. Linguraru
Graph-Based Deep Learning on Stereo EEG for Predicting Seizure Freedom in Epilepsy Patients
Feb 21, 2025Abstract:Predicting seizure freedom is essential for tailoring epilepsy treatment. But accurate prediction remains challenging with traditional methods, especially with diverse patient populations. This study developed a deep learning-based graph neural network (GNN) model to predict seizure freedom from stereo electroencephalography (sEEG) data in patients with refractory epilepsy. We utilized high-quality sEEG data from 15 pediatric patients to train a deep learning model that can accurately predict seizure freedom outcomes and advance understanding of brain connectivity at the seizure onset zone. Our model integrates local and global connectivity using graph convolutions with multi-scale attention mechanisms to capture connections between difficult-to-study regions such as the thalamus and motor regions. The model achieved an accuracy of 92.4% in binary class analysis, 86.6% in patient-wise analysis, and 81.4% in multi-class analysis. Node and edge-level feature analysis highlighted the anterior cingulate and frontal pole regions as key contributors to seizure freedom outcomes. The nodes identified by our model were also more likely to coincide with seizure onset zones. Our findings underscore the potential of new connectivity-based deep learning models such as GNNs for enhancing the prediction of seizure freedom, predicting seizure onset zones, connectivity analysis of the brain during seizure, as well as informing AI-assisted personalized epilepsy treatment planning.
A New Logic For Pediatric Brain Tumor Segmentation
Nov 03, 2024



Abstract:In this paper, we present a novel approach for segmenting pediatric brain tumors using a deep learning architecture, inspired by expert radiologists' segmentation strategies. Our model delineates four distinct tumor labels and is benchmarked on a held-out PED BraTS 2024 test set (i.e., pediatric brain tumor datasets introduced by BraTS). Furthermore, we evaluate our model's performance against the state-of-the-art (SOTA) model using a new external dataset of 30 patients from CBTN (Children's Brain Tumor Network), labeled in accordance with the PED BraTS 2024 guidelines. We compare segmentation outcomes with the winning algorithm from the PED BraTS 2023 challenge as the SOTA model. Our proposed algorithm achieved an average Dice score of 0.642 and an HD95 of 73.0 mm on the CBTN test data, outperforming the SOTA model, which achieved a Dice score of 0.626 and an HD95 of 84.0 mm. Our results indicate that the proposed model is a step towards providing more accurate segmentation for pediatric brain tumors, which is essential for evaluating therapy response and monitoring patient progress.
Zero-Shot Pediatric Tuberculosis Detection in Chest X-Rays using Self-Supervised Learning
Feb 22, 2024



Abstract:Tuberculosis (TB) remains a significant global health challenge, with pediatric cases posing a major concern. The World Health Organization (WHO) advocates for chest X-rays (CXRs) for TB screening. However, visual interpretation by radiologists can be subjective, time-consuming and prone to error, especially in pediatric TB. Artificial intelligence (AI)-driven computer-aided detection (CAD) tools, especially those utilizing deep learning, show promise in enhancing lung disease detection. However, challenges include data scarcity and lack of generalizability. In this context, we propose a novel self-supervised paradigm leveraging Vision Transformers (ViT) for improved TB detection in CXR, enabling zero-shot pediatric TB detection. We demonstrate improvements in TB detection performance ($\sim$12.7% and $\sim$13.4% top AUC/AUPR gains in adults and children, respectively) when conducting self-supervised pre-training when compared to fully-supervised (i.e., non pre-trained) ViT models, achieving top performances of 0.959 AUC and 0.962 AUPR in adult TB detection, and 0.697 AUC and 0.607 AUPR in zero-shot pediatric TB detection. As a result, this work demonstrates that self-supervised learning on adult CXRs effectively extends to challenging downstream tasks such as pediatric TB detection, where data are scarce.
DiCoM -- Diverse Concept Modeling towards Enhancing Generalizability in Chest X-Ray Studies
Feb 22, 2024Abstract:Chest X-Ray (CXR) is a widely used clinical imaging modality and has a pivotal role in the diagnosis and prognosis of various lung and heart related conditions. Conventional automated clinical diagnostic tool design strategies relying on radiology reads and supervised learning, entail the cumbersome requirement of high quality annotated training data. To address this challenge, self-supervised pre-training has proven to outperform supervised pre-training in numerous downstream vision tasks, representing a significant breakthrough in the field. However, medical imaging pre-training significantly differs from pre-training with natural images (e.g., ImageNet) due to unique attributes of clinical images. In this context, we introduce Diverse Concept Modeling (DiCoM), a novel self-supervised training paradigm that leverages a student teacher framework for learning diverse concepts and hence effective representation of the CXR data. Hence, expanding beyond merely modeling a single primary label within an image, instead, effectively harnessing the information from all the concepts inherent in the CXR. The pre-trained model is subsequently fine-tuned to address diverse domain-specific tasks. Our proposed paradigm consistently demonstrates robust performance across multiple downstream tasks on multiple datasets, highlighting the success and generalizability of the pre-training strategy. To establish the efficacy of our methods we analyze both the power of learned representations and the speed of convergence (SoC) of our models. For diverse data and tasks, DiCoM is able to achieve in most cases better results compared to other state-of-the-art pre-training strategies. This when combined with the higher SoC and generalization capabilities positions DiCoM to be established as a foundation model for CXRs, a widely used imaging modality.
Quantitative Metrics for Benchmarking Medical Image Harmonization
Feb 06, 2024



Abstract:Image harmonization is an important preprocessing strategy to address domain shifts arising from data acquired using different machines and scanning protocols in medical imaging. However, benchmarking the effectiveness of harmonization techniques has been a challenge due to the lack of widely available standardized datasets with ground truths. In this context, we propose three metrics: two intensity harmonization metrics and one anatomy preservation metric for medical images during harmonization, where no ground truths are required. Through extensive studies on a dataset with available harmonization ground truth, we demonstrate that our metrics are correlated with established image quality assessment metrics. We show how these novel metrics may be applied to real-world scenarios where no harmonization ground truth exists. Additionally, we provide insights into different interpretations of the metric values, shedding light on their significance in the context of the harmonization process. As a result of our findings, we advocate for the adoption of these quantitative harmonization metrics as a standard for benchmarking the performance of image harmonization techniques.
Partitioned Shape Modeling with On-the-Fly Sparse Appearance Learning for Anterior Visual Pathway Segmentation
Aug 05, 2015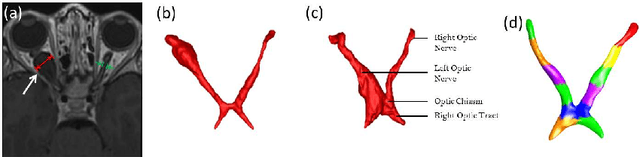
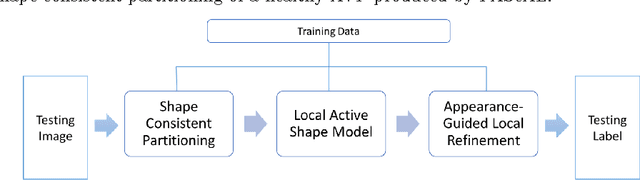
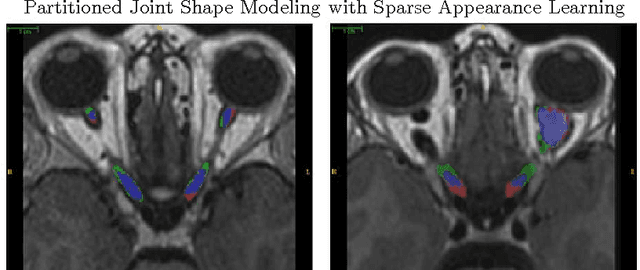
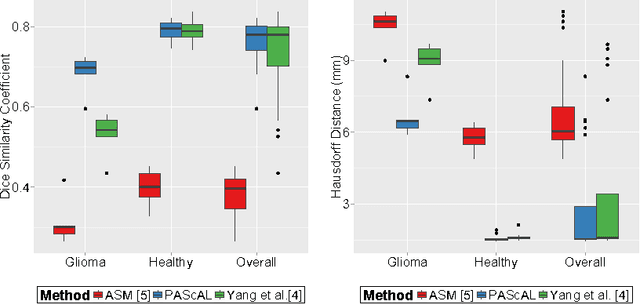
Abstract:MRI quantification of cranial nerves such as anterior visual pathway (AVP) in MRI is challenging due to their thin small size, structural variation along its path, and adjacent anatomic structures. Segmentation of pathologically abnormal optic nerve (e.g. optic nerve glioma) poses additional challenges due to changes in its shape at unpredictable locations. In this work, we propose a partitioned joint statistical shape model approach with sparse appearance learning for the segmentation of healthy and pathological AVP. Our main contributions are: (1) optimally partitioned statistical shape models for the AVP based on regional shape variations for greater local flexibility of statistical shape model; (2) refinement model to accommodate pathological regions as well as areas of subtle variation by training the model on-the-fly using the initial segmentation obtained in (1); (3) hierarchical deformable framework to incorporate scale information in partitioned shape and appearance models. Our method, entitled PAScAL (PArtitioned Shape and Appearance Learning), was evaluated on 21 MRI scans (15 healthy + 6 glioma cases) from pediatric patients (ages 2-17). The experimental results show that the proposed localized shape and sparse appearance-based learning approach significantly outperforms segmentation approaches in the analysis of pathological data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge