Awais Mansoor
Generation of Radiology Findings in Chest X-Ray by Leveraging Collaborative Knowledge
Jun 18, 2023



Abstract:Among all the sub-sections in a typical radiology report, the Clinical Indications, Findings, and Impression often reflect important details about the health status of a patient. The information included in Impression is also often covered in Findings. While Findings and Impression can be deduced by inspecting the image, Clinical Indications often require additional context. The cognitive task of interpreting medical images remains the most critical and often time-consuming step in the radiology workflow. Instead of generating an end-to-end radiology report, in this paper, we focus on generating the Findings from automated interpretation of medical images, specifically chest X-rays (CXRs). Thus, this work focuses on reducing the workload of radiologists who spend most of their time either writing or narrating the Findings. Unlike past research, which addresses radiology report generation as a single-step image captioning task, we have further taken into consideration the complexity of interpreting CXR images and propose a two-step approach: (a) detecting the regions with abnormalities in the image, and (b) generating relevant text for regions with abnormalities by employing a generative large language model (LLM). This two-step approach introduces a layer of interpretability and aligns the framework with the systematic reasoning that radiologists use when reviewing a CXR.
* Information Technology and Quantitative Management (ITQM 2023)
Self-supervised Learning from 100 Million Medical Images
Jan 04, 2022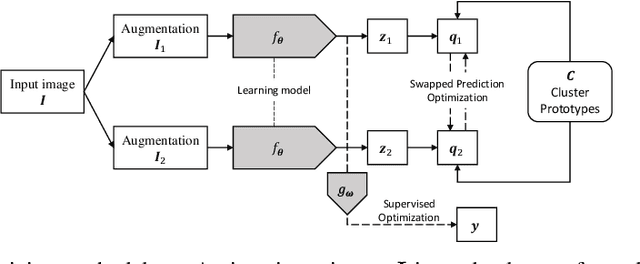
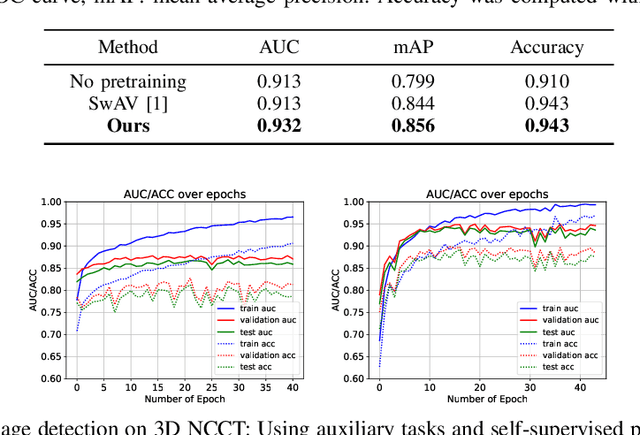
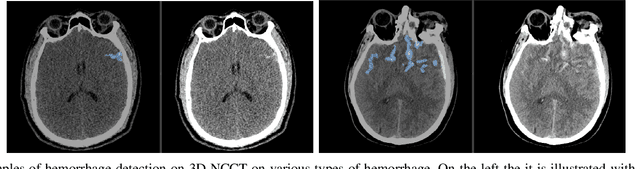
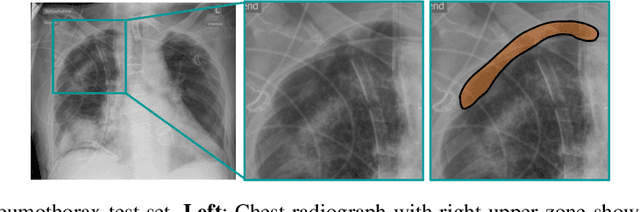
Abstract:Building accurate and robust artificial intelligence systems for medical image assessment requires not only the research and design of advanced deep learning models but also the creation of large and curated sets of annotated training examples. Constructing such datasets, however, is often very costly -- due to the complex nature of annotation tasks and the high level of expertise required for the interpretation of medical images (e.g., expert radiologists). To counter this limitation, we propose a method for self-supervised learning of rich image features based on contrastive learning and online feature clustering. For this purpose we leverage large training datasets of over 100,000,000 medical images of various modalities, including radiography, computed tomography (CT), magnetic resonance (MR) imaging and ultrasonography. We propose to use these features to guide model training in supervised and hybrid self-supervised/supervised regime on various downstream tasks. We highlight a number of advantages of this strategy on challenging image assessment problems in radiography, CT and MR: 1) Significant increase in accuracy compared to the state-of-the-art (e.g., AUC boost of 3-7% for detection of abnormalities from chest radiography scans and hemorrhage detection on brain CT); 2) Acceleration of model convergence during training by up to 85% compared to using no pretraining (e.g., 83% when training a model for detection of brain metastases in MR scans); 3) Increase in robustness to various image augmentations, such as intensity variations, rotations or scaling reflective of data variation seen in the field.
Automated detection and quantification of COVID-19 airspace disease on chest radiographs: A novel approach achieving radiologist-level performance using a CNN trained on digital reconstructed radiographs (DRRs) from CT-based ground-truth
Aug 13, 2020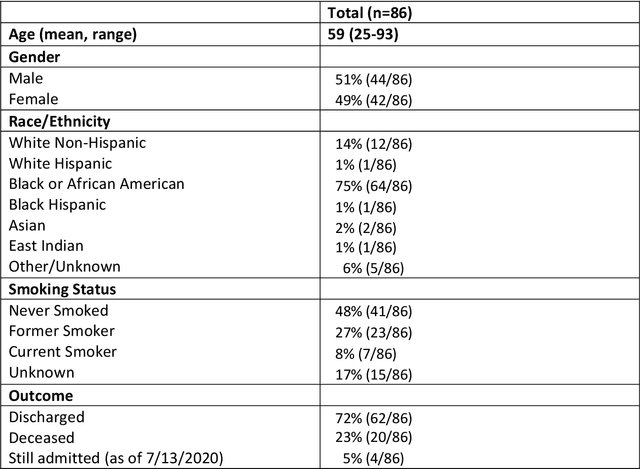
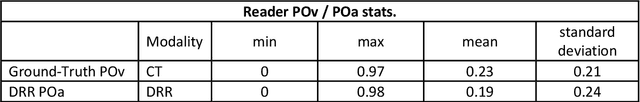
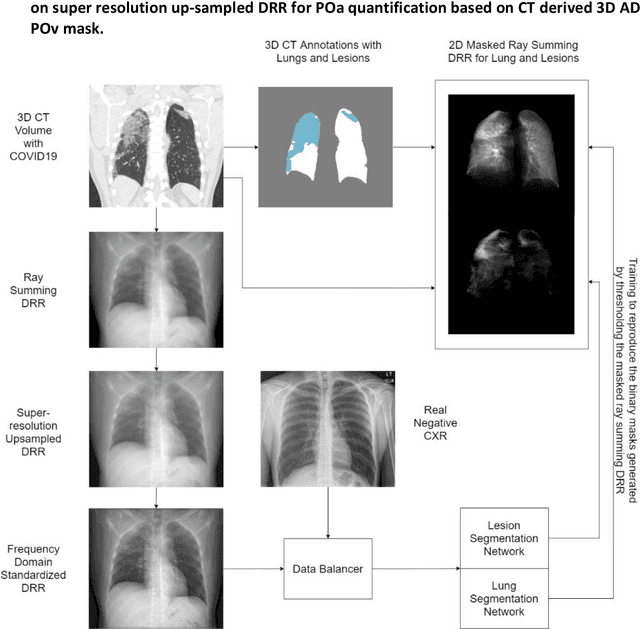
Abstract:Purpose: To leverage volumetric quantification of airspace disease (AD) derived from a superior modality (CT) serving as ground truth, projected onto digitally reconstructed radiographs (DRRs) to: 1) train a convolutional neural network to quantify airspace disease on paired CXRs; and 2) compare the DRR-trained CNN to expert human readers in the CXR evaluation of patients with confirmed COVID-19. Materials and Methods: We retrospectively selected a cohort of 86 COVID-19 patients (with positive RT-PCR), from March-May 2020 at a tertiary hospital in the northeastern USA, who underwent chest CT and CXR within 48 hrs. The ground truth volumetric percentage of COVID-19 related AD (POv) was established by manual AD segmentation on CT. The resulting 3D masks were projected into 2D anterior-posterior digitally reconstructed radiographs (DRR) to compute area-based AD percentage (POa). A convolutional neural network (CNN) was trained with DRR images generated from a larger-scale CT dataset of COVID-19 and non-COVID-19 patients, automatically segmenting lungs, AD and quantifying POa on CXR. CNN POa results were compared to POa quantified on CXR by two expert readers and to the POv ground-truth, by computing correlations and mean absolute errors. Results: Bootstrap mean absolute error (MAE) and correlations between POa and POv were 11.98% [11.05%-12.47%] and 0.77 [0.70-0.82] for average of expert readers, and 9.56%-9.78% [8.83%-10.22%] and 0.78-0.81 [0.73-0.85] for the CNN, respectively. Conclusion: Our CNN trained with DRR using CT-derived airspace quantification achieved expert radiologist level of accuracy in the quantification of airspace disease on CXR, in patients with positive RT-PCR for COVID-19.
Quantifying and Leveraging Predictive Uncertainty for Medical Image Assessment
Jul 08, 2020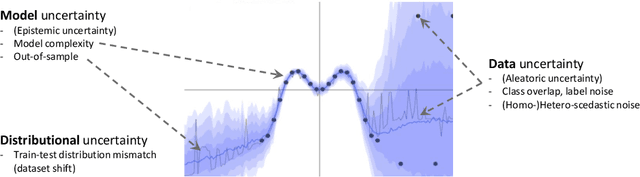

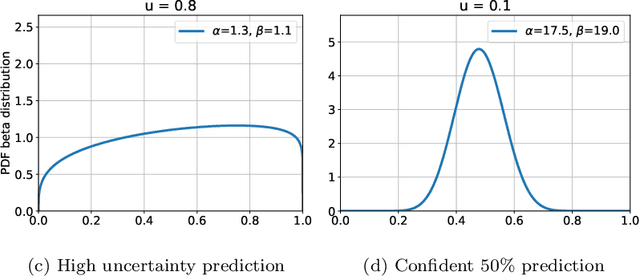
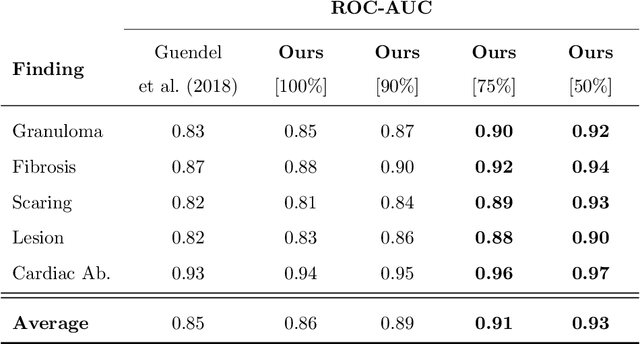
Abstract:The interpretation of medical images is a challenging task, often complicated by the presence of artifacts, occlusions, limited contrast and more. Most notable is the case of chest radiography, where there is a high inter-rater variability in the detection and classification of abnormalities. This is largely due to inconclusive evidence in the data or subjective definitions of disease appearance. An additional example is the classification of anatomical views based on 2D Ultrasound images. Often, the anatomical context captured in a frame is not sufficient to recognize the underlying anatomy. Current machine learning solutions for these problems are typically limited to providing probabilistic predictions, relying on the capacity of underlying models to adapt to limited information and the high degree of label noise. In practice, however, this leads to overconfident systems with poor generalization on unseen data. To account for this, we propose a system that learns not only the probabilistic estimate for classification, but also an explicit uncertainty measure which captures the confidence of the system in the predicted output. We argue that this approach is essential to account for the inherent ambiguity characteristic of medical images from different radiologic exams including computed radiography, ultrasonography and magnetic resonance imaging. In our experiments we demonstrate that sample rejection based on the predicted uncertainty can significantly improve the ROC-AUC for various tasks, e.g., by 8% to 0.91 with an expected rejection rate of under 25% for the classification of different abnormalities in chest radiographs. In addition, we show that using uncertainty-driven bootstrapping to filter the training data, one can achieve a significant increase in robustness and accuracy.
Communal Domain Learning for Registration in Drifted Image Spaces
Aug 20, 2019



Abstract:Designing a registration framework for images that do not share the same probability distribution is a major challenge in modern image analytics yet trivial task for the human visual system (HVS). Discrepancies in probability distributions, also known as \emph{drifts}, can occur due to various reasons including, but not limited to differences in sequences and modalities (e.g., MRI T1-T2 and MRI-CT registration), or acquisition settings (e.g., multisite, inter-subject, or intra-subject registrations). The popular assumption about the working of HVS is that it exploits a communal feature subspace exists between the registering images or fields-of-view that encompasses key drift-invariant features. Mimicking the approach that is potentially adopted by the HVS, herein, we present a representation learning technique of this invariant communal subspace that is shared by registering domains. The proposed communal domain learning (CDL) framework uses a set of hierarchical nonlinear transforms to learn the communal subspace that minimizes the probability differences and maximizes the amount of shared information between the registering domains. Similarity metric and parameter optimization calculations for registration are subsequently performed in the drift-minimized learned communal subspace. This generic registration framework is applied to register multisequence (MR: T1, T2) and multimodal (MR, CT) images. Results demonstrated generic applicability, consistent performance, and statistically significant improvement for both multi-sequence and multi-modal data using the proposed approach ($p$-value$<0.001$; Wilcoxon rank sum test) over baseline methods.
Region Proposal Networks with Contextual Selective Attention for Real-Time Organ Detection
Dec 26, 2018


Abstract:State-of-the-art methods for object detection use region proposal networks (RPN) to hypothesize object location. These networks simultaneously predicts object bounding boxes and \emph{objectness} scores at each location in the image. Unlike natural images for which RPN algorithms were originally designed, most medical images are acquired following standard protocols, thus organs in the image are typically at a similar location and possess similar geometrical characteristics (e.g. scale, aspect-ratio, etc.). Therefore, medical image acquisition protocols hold critical localization and geometric information that can be incorporated for faster and more accurate detection. This paper presents a novel attention mechanism for the detection of organs by incorporating imaging protocol information. Our novel selective attention approach (i) effectively shrinks the search space inside the feature map, (ii) appends useful localization information to the hypothesized proposal for the detection architecture to learn where to look for each organ, and (iii) modifies the pyramid of regression references in the RPN by incorporating organ- and modality-specific information, which results in additional time reduction. We evaluated the proposed framework on a dataset of 768 chest X-ray images obtained from a diverse set of sources. Our results demonstrate superior performance for the detection of the lung field compared to the state-of-the-art, both in terms of detection accuracy, demonstrating an improvement of $>7\%$ in Dice score, and reduced processing time by $27.53\%$ due to fewer hypotheses.
A Generic Approach to Lung Field Segmentation from Chest Radiographs using Deep Space and Shape Learning
Jul 11, 2018



Abstract:Computer-aided diagnosis (CAD) techniques for lung field segmentation from chest radiographs (CXR) have been proposed for adult cohorts, but rarely for pediatric subjects. Statistical shape models (SSMs), the workhorse of most state-of-the-art CXR-based lung field segmentation methods, do not efficiently accommodate shape variation of the lung field during the pediatric developmental stages. The main contributions of our work are: (1) a generic lung field segmentation framework from CXR accommodating large shape variation for adult and pediatric cohorts; (2) a deep representation learning detection mechanism, \emph{ensemble space learning}, for robust object localization; and (3) \emph{marginal shape deep learning} for the shape deformation parameter estimation. Unlike the iterative approach of conventional SSMs, the proposed shape learning mechanism transforms the parameter space into marginal subspaces that are solvable efficiently using the recursive representation learning mechanism. Furthermore, our method is the first to include the challenging retro-cardiac region in the CXR-based lung segmentation for accurate lung capacity estimation. The framework is evaluated on 668 CXRs of patients between 3 month to 89 year of age. We obtain a mean Dice similarity coefficient of $0.96\pm0.03$ (including the retro-cardiac region). For a given accuracy, the proposed approach is also found to be faster than conventional SSM-based iterative segmentation methods. The computational simplicity of the proposed generic framework could be similarly applied to the fast segmentation of other deformable objects.
Partitioned Shape Modeling with On-the-Fly Sparse Appearance Learning for Anterior Visual Pathway Segmentation
Aug 05, 2015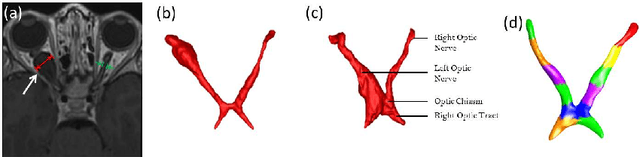
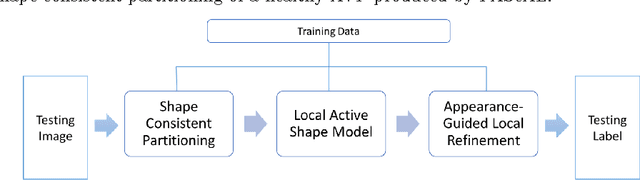
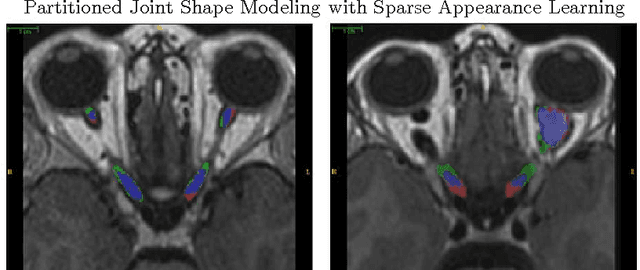
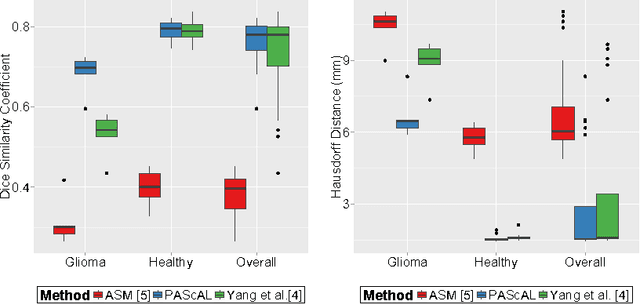
Abstract:MRI quantification of cranial nerves such as anterior visual pathway (AVP) in MRI is challenging due to their thin small size, structural variation along its path, and adjacent anatomic structures. Segmentation of pathologically abnormal optic nerve (e.g. optic nerve glioma) poses additional challenges due to changes in its shape at unpredictable locations. In this work, we propose a partitioned joint statistical shape model approach with sparse appearance learning for the segmentation of healthy and pathological AVP. Our main contributions are: (1) optimally partitioned statistical shape models for the AVP based on regional shape variations for greater local flexibility of statistical shape model; (2) refinement model to accommodate pathological regions as well as areas of subtle variation by training the model on-the-fly using the initial segmentation obtained in (1); (3) hierarchical deformable framework to incorporate scale information in partitioned shape and appearance models. Our method, entitled PAScAL (PArtitioned Shape and Appearance Learning), was evaluated on 21 MRI scans (15 healthy + 6 glioma cases) from pediatric patients (ages 2-17). The experimental results show that the proposed localized shape and sparse appearance-based learning approach significantly outperforms segmentation approaches in the analysis of pathological data.
Optimally Stabilized PET Image Denoising Using Trilateral Filtering
Jul 11, 2014



Abstract:Low-resolution and signal-dependent noise distribution in positron emission tomography (PET) images makes denoising process an inevitable step prior to qualitative and quantitative image analysis tasks. Conventional PET denoising methods either over-smooth small-sized structures due to resolution limitation or make incorrect assumptions about the noise characteristics. Therefore, clinically important quantitative information may be corrupted. To address these challenges, we introduced a novel approach to remove signal-dependent noise in the PET images where the noise distribution was considered as Poisson-Gaussian mixed. Meanwhile, the generalized Anscombe's transformation (GAT) was used to stabilize varying nature of the PET noise. Other than noise stabilization, it is also desirable for the noise removal filter to preserve the boundaries of the structures while smoothing the noisy regions. Indeed, it is important to avoid significant loss of quantitative information such as standard uptake value (SUV)-based metrics as well as metabolic lesion volume. To satisfy all these properties, we extended bilateral filtering method into trilateral filtering through multiscaling and optimal Gaussianization process. The proposed method was tested on more than 50 PET-CT images from various patients having different cancers and achieved the superior performance compared to the widely used denoising techniques in the literature.
Near-optimal Keypoint Sampling for Fast Pathological Lung Segmentation
Jul 11, 2014



Abstract:Accurate delineation of pathological lungs from computed tomography (CT) images remains mostly unsolved because available methods fail to provide a reliable generic solution due to high variability of abnormality appearance. Local descriptor-based classification methods have shown to work well in annotating pathologies; however, these methods are usually computationally intensive which restricts their widespread use in real-time or near-real-time clinical applications. In this paper, we present a novel approach for fast, accurate, reliable segmentation of pathological lungs from CT scans by combining region-based segmentation method with local descriptor classification that is performed on an optimized sampling grid. Our method works in two stages; during stage one, we adapted the fuzzy connectedness (FC) image segmentation algorithm to perform initial lung parenchyma extraction. In the second stage, texture-based local descriptors are utilized to segment abnormal imaging patterns using a near optimal keypoint analysis by employing centroid of supervoxel as grid points. The quantitative results show that our pathological lung segmentation method is fast, robust, and improves on current standards and has potential to enhance the performance of routine clinical tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge