Florin C. Ghesu
A Beam's Eye View to Fluence Maps 3D Network for Ultra Fast VMAT Radiotherapy Planning
Feb 05, 2025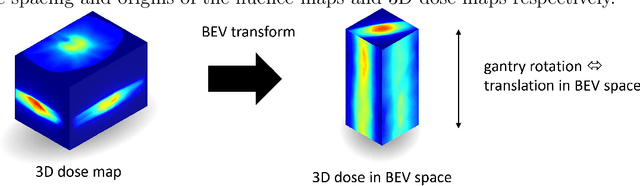
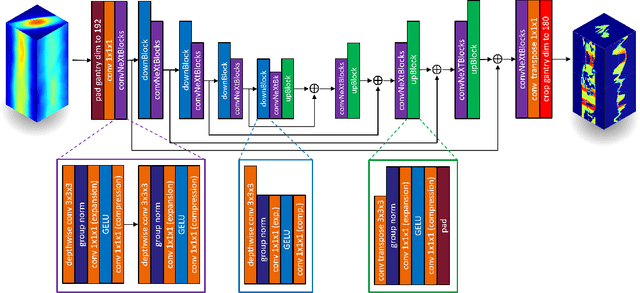

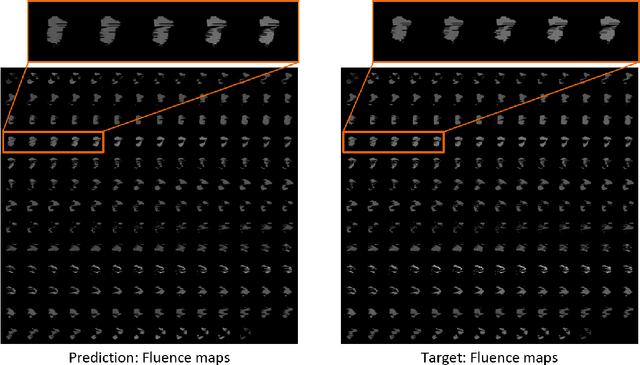
Abstract:Volumetric Modulated Arc Therapy (VMAT) revolutionizes cancer treatment by precisely delivering radiation while sparing healthy tissues. Fluence maps generation, crucial in VMAT planning, traditionally involves complex and iterative, and thus time consuming processes. These fluence maps are subsequently leveraged for leaf-sequence. The deep-learning approach presented in this article aims to expedite this by directly predicting fluence maps from patient data. We developed a 3D network which we trained in a supervised way using a combination of L1 and L2 losses, and RT plans generated by Eclipse and from the REQUITE dataset, taking the RT dose map as input and the fluence maps computed from the corresponding RT plans as target. Our network predicts jointly the 180 fluence maps corresponding to the 180 control points (CP) of single arc VMAT plans. In order to help the network, we pre-process the input dose by computing the projections of the 3D dose map to the beam's eye view (BEV) of the 180 CPs, in the same coordinate system as the fluence maps. We generated over 2000 VMAT plans using Eclipse to scale up the dataset size. Additionally, we evaluated various network architectures and analyzed the impact of increasing the dataset size. We are measuring the performance in the 2D fluence maps domain using image metrics (PSNR, SSIM), as well as in the 3D dose domain using the dose-volume histogram (DVH) on a validation dataset. The network inference, which does not include the data loading and processing, is less than 20ms. Using our proposed 3D network architecture as well as increasing the dataset size using Eclipse improved the fluence map reconstruction performance by approximately 8 dB in PSNR compared to a U-Net architecture trained on the original REQUITE dataset. The resulting DVHs are very close to the one of the input target dose.
Automating High Quality RT Planning at Scale
Jan 21, 2025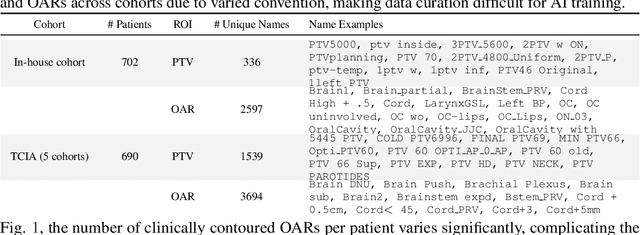
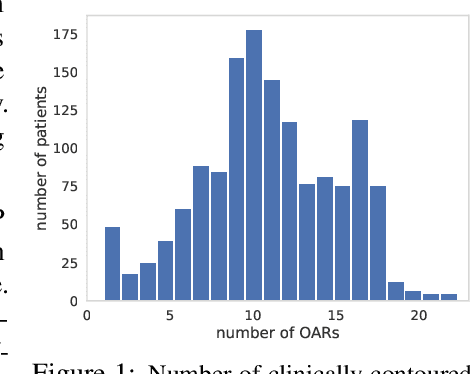
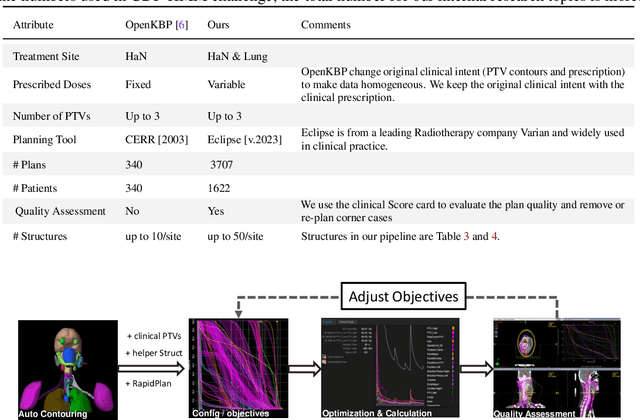
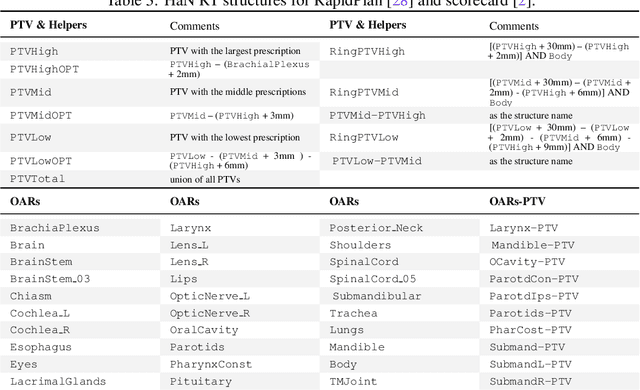
Abstract:Radiotherapy (RT) planning is complex, subjective, and time-intensive. Advances in artificial intelligence (AI) promise to improve its precision, efficiency, and consistency, but progress is often limited by the scarcity of large, standardized datasets. To address this, we introduce the Automated Iterative RT Planning (AIRTP) system, a scalable solution for generating high-quality treatment plans. This scalable solution is designed to generate substantial volumes of consistently high-quality treatment plans, overcoming a key obstacle in the advancement of AI-driven RT planning. Our AIRTP pipeline adheres to clinical guidelines and automates essential steps, including organ-at-risk (OAR) contouring, helper structure creation, beam setup, optimization, and plan quality improvement, using AI integrated with RT planning software like Eclipse of Varian. Furthermore, a novel approach for determining optimization parameters to reproduce 3D dose distributions, i.e. a method to convert dose predictions to deliverable treatment plans constrained by machine limitations. A comparative analysis of plan quality reveals that our automated pipeline produces treatment plans of quality comparable to those generated manually, which traditionally require several hours of labor per plan. Committed to public research, the first data release of our AIRTP pipeline includes nine cohorts covering head-and-neck and lung cancer sites to support an AAPM 2025 challenge. This data set features more than 10 times the number of plans compared to the largest existing well-curated public data set to our best knowledge. Repo:{https://github.com/RiqiangGao/GDP-HMM_AAPMChallenge}
Towards Integrating Epistemic Uncertainty Estimation into the Radiotherapy Workflow
Sep 27, 2024Abstract:The precision of contouring target structures and organs-at-risk (OAR) in radiotherapy planning is crucial for ensuring treatment efficacy and patient safety. Recent advancements in deep learning (DL) have significantly improved OAR contouring performance, yet the reliability of these models, especially in the presence of out-of-distribution (OOD) scenarios, remains a concern in clinical settings. This application study explores the integration of epistemic uncertainty estimation within the OAR contouring workflow to enable OOD detection in clinically relevant scenarios, using specifically compiled data. Furthermore, we introduce an advanced statistical method for OOD detection to enhance the methodological framework of uncertainty estimation. Our empirical evaluation demonstrates that epistemic uncertainty estimation is effective in identifying instances where model predictions are unreliable and may require an expert review. Notably, our approach achieves an AUC-ROC of 0.95 for OOD detection, with a specificity of 0.95 and a sensitivity of 0.92 for implant cases, underscoring its efficacy. This study addresses significant gaps in the current research landscape, such as the lack of ground truth for uncertainty estimation and limited empirical evaluations. Additionally, it provides a clinically relevant application of epistemic uncertainty estimation in an FDA-approved and widely used clinical solution for OAR segmentation from Varian, a Siemens Healthineers company, highlighting its practical benefits.
Multi-Agent Reinforcement Learning Meets Leaf Sequencing in Radiotherapy
Jun 03, 2024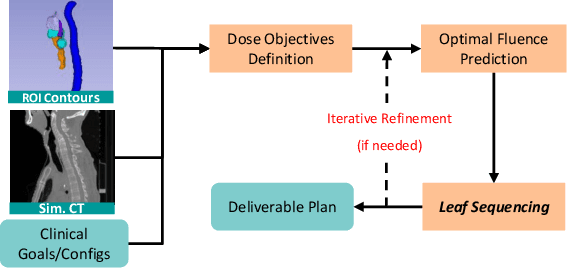
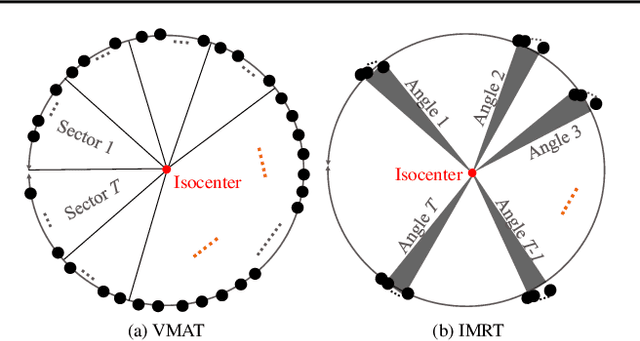
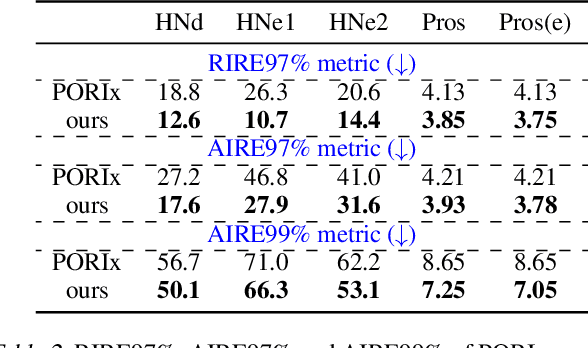
Abstract:In contemporary radiotherapy planning (RTP), a key module leaf sequencing is predominantly addressed by optimization-based approaches. In this paper, we propose a novel deep reinforcement learning (DRL) model termed as Reinforced Leaf Sequencer (RLS) in a multi-agent framework for leaf sequencing. The RLS model offers improvements to time-consuming iterative optimization steps via large-scale training and can control movement patterns through the design of reward mechanisms. We have conducted experiments on four datasets with four metrics and compared our model with a leading optimization sequencer. Our findings reveal that the proposed RLS model can achieve reduced fluence reconstruction errors, and potential faster convergence when integrated in an optimization planner. Additionally, RLS has shown promising results in a full artificial intelligence RTP pipeline. We hope this pioneer multi-agent RL leaf sequencer can foster future research on machine learning for RTP.
Goal-conditioned reinforcement learning for ultrasound navigation guidance
May 02, 2024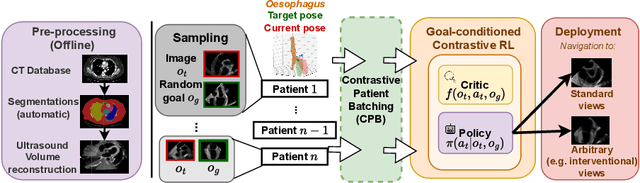
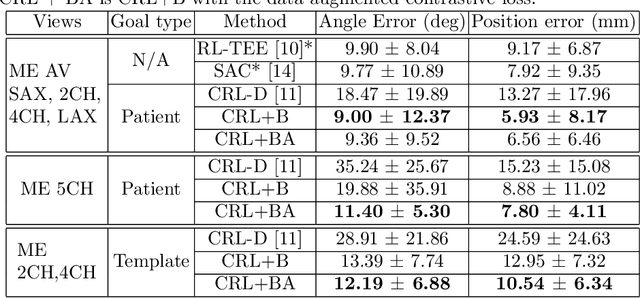
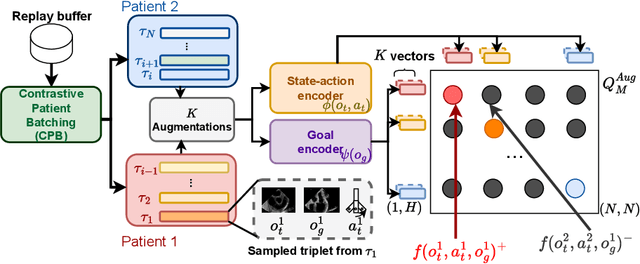
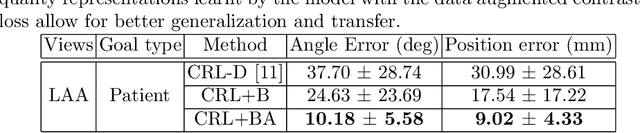
Abstract:Transesophageal echocardiography (TEE) plays a pivotal role in cardiology for diagnostic and interventional procedures. However, using it effectively requires extensive training due to the intricate nature of image acquisition and interpretation. To enhance the efficiency of novice sonographers and reduce variability in scan acquisitions, we propose a novel ultrasound (US) navigation assistance method based on contrastive learning as goal-conditioned reinforcement learning (GCRL). We augment the previous framework using a novel contrastive patient batching method (CPB) and a data-augmented contrastive loss, both of which we demonstrate are essential to ensure generalization to anatomical variations across patients. The proposed framework enables navigation to both standard diagnostic as well as intricate interventional views with a single model. Our method was developed with a large dataset of 789 patients and obtained an average error of 6.56 mm in position and 9.36 degrees in angle on a testing dataset of 140 patients, which is competitive or superior to models trained on individual views. Furthermore, we quantitatively validate our method's ability to navigate to interventional views such as the Left Atrial Appendage (LAA) view used in LAA closure. Our approach holds promise in providing valuable guidance during transesophageal ultrasound examinations, contributing to the advancement of skill acquisition for cardiac ultrasound practitioners.
Self-Supervised Learning for Interventional Image Analytics: Towards Robust Device Trackers
May 02, 2024Abstract:An accurate detection and tracking of devices such as guiding catheters in live X-ray image acquisitions is an essential prerequisite for endovascular cardiac interventions. This information is leveraged for procedural guidance, e.g., directing stent placements. To ensure procedural safety and efficacy, there is a need for high robustness no failures during tracking. To achieve that, one needs to efficiently tackle challenges, such as: device obscuration by contrast agent or other external devices or wires, changes in field-of-view or acquisition angle, as well as the continuous movement due to cardiac and respiratory motion. To overcome the aforementioned challenges, we propose a novel approach to learn spatio-temporal features from a very large data cohort of over 16 million interventional X-ray frames using self-supervision for image sequence data. Our approach is based on a masked image modeling technique that leverages frame interpolation based reconstruction to learn fine inter-frame temporal correspondences. The features encoded in the resulting model are fine-tuned downstream. Our approach achieves state-of-the-art performance and in particular robustness compared to ultra optimized reference solutions (that use multi-stage feature fusion, multi-task and flow regularization). The experiments show that our method achieves 66.31% reduction in maximum tracking error against reference solutions (23.20% when flow regularization is used); achieving a success score of 97.95% at a 3x faster inference speed of 42 frames-per-second (on GPU). The results encourage the use of our approach in various other tasks within interventional image analytics that require effective understanding of spatio-temporal semantics.
ConTrack: Contextual Transformer for Device Tracking in X-ray
Jul 14, 2023Abstract:Device tracking is an important prerequisite for guidance during endovascular procedures. Especially during cardiac interventions, detection and tracking of guiding the catheter tip in 2D fluoroscopic images is important for applications such as mapping vessels from angiography (high dose with contrast) to fluoroscopy (low dose without contrast). Tracking the catheter tip poses different challenges: the tip can be occluded by contrast during angiography or interventional devices; and it is always in continuous movement due to the cardiac and respiratory motions. To overcome these challenges, we propose ConTrack, a transformer-based network that uses both spatial and temporal contextual information for accurate device detection and tracking in both X-ray fluoroscopy and angiography. The spatial information comes from the template frames and the segmentation module: the template frames define the surroundings of the device, whereas the segmentation module detects the entire device to bring more context for the tip prediction. Using multiple templates makes the model more robust to the change in appearance of the device when it is occluded by the contrast agent. The flow information computed on the segmented catheter mask between the current and the previous frame helps in further refining the prediction by compensating for the respiratory and cardiac motions. The experiments show that our method achieves 45% or higher accuracy in detection and tracking when compared to state-of-the-art tracking models.
AI-based Agents for Automated Robotic Endovascular Guidewire Manipulation
Apr 18, 2023Abstract:Endovascular guidewire manipulation is essential for minimally-invasive clinical applications (Percutaneous Coronary Intervention (PCI), Mechanical thrombectomy techniques for acute ischemic stroke (AIS), or Transjugular intrahepatic portosystemic shunt (TIPS)). All procedures commonly require 3D vessel geometries from 3D CTA (Computed Tomography Angiography) images. During these procedures, the clinician generally places a guiding catheter in the ostium of the relevant vessel and then manipulates a wire through the catheter and across the blockage. The clinician only uses X-ray fluoroscopy intermittently to visualize and guide the catheter, guidewire, and other devices. However, clinicians still passively control guidewires/catheters by relying on limited indirect observation (i.e., 2D partial view of devices, and intermittent updates due to radiation limit) from X-ray fluoroscopy. Modeling and controlling the guidewire manipulation in coronary vessels remains challenging because of the complicated interaction between guidewire motions with different physical properties (i.e., loads, coating) and vessel geometries with lumen conditions resulting in a highly non-linear system. This paper introduces a scalable learning pipeline to train AI-based agent models toward automated endovascular predictive device controls. First, we create a scalable environment by pre-processing 3D CTA images, providing patient-specific 3D vessel geometry and the centerline of the coronary. Next, we apply a large quantity of randomly generated motion sequences from the proximal end to generate wire states associated with each environment using a physics-based device simulator. Then, we reformulate the control problem to a sequence-to-sequence learning problem, in which we use a Transformer-based model, trained to handle non-linear sequential forward/inverse transition functions.
Design, Modeling, and Evaluation of Separable Tendon-Driven Robotic Manipulator with Long, Passive, Flexible Proximal Section
Jan 01, 2023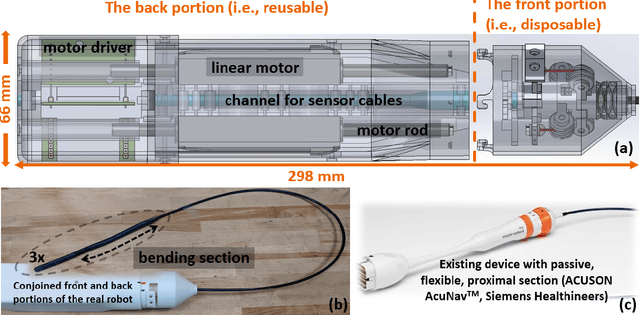

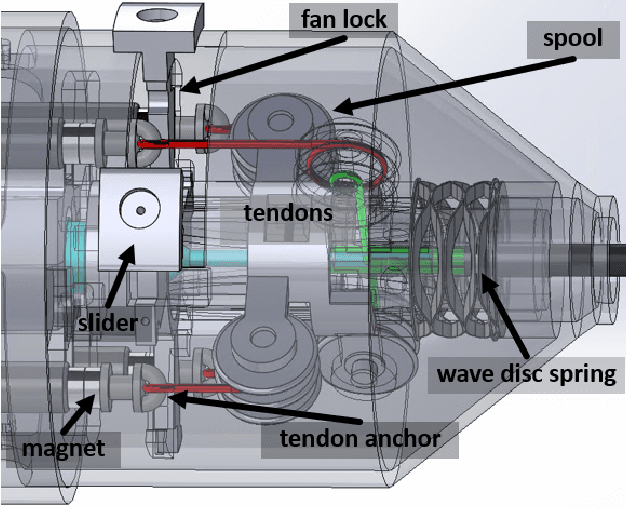

Abstract:The purpose of this work was to tackle practical issues which arise when using a tendon-driven robotic manipulator with a long, passive, flexible proximal section in medical applications. A separable robot which overcomes difficulties in actuation and sterilization is introduced, in which the body containing the electronics is reusable and the remainder is disposable. A control input which resolves the redundancy in the kinematics and a physical interpretation of this redundancy are provided. The effect of a static change in the proximal section angle on bending angle error was explored under four testing conditions for a sinusoidal input. Bending angle error increased for increasing proximal section angle for all testing conditions with an average error reduction of 41.48% for retension, 4.28% for hysteresis, and 52.35% for re-tension + hysteresis compensation relative to the baseline case. Two major sources of error in tracking the bending angle were identified: time delay from hysteresis and DC offset from the proximal section angle. Examination of these error sources revealed that the simple hysteresis compensation was most effective for removing time delay and re-tension compensation for removing DC offset, which was the primary source of increasing error. The re-tension compensation was also tested for dynamic changes in the proximal section and reduced error in the final configuration of the tip by 89.14% relative to the baseline case.
Self-supervised Learning from 100 Million Medical Images
Jan 04, 2022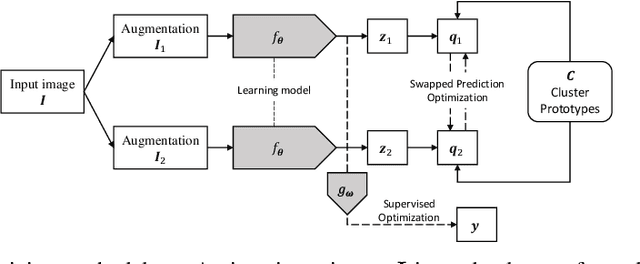
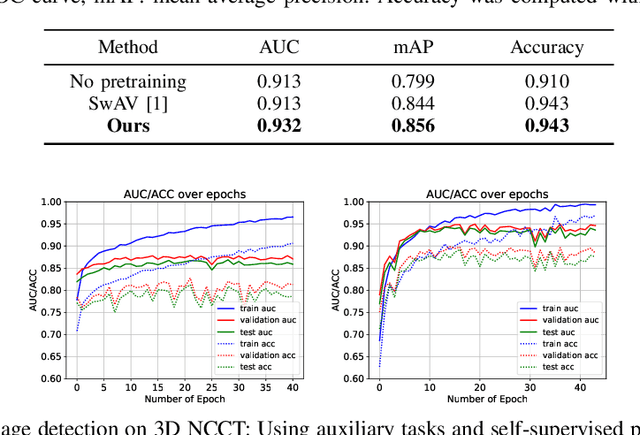
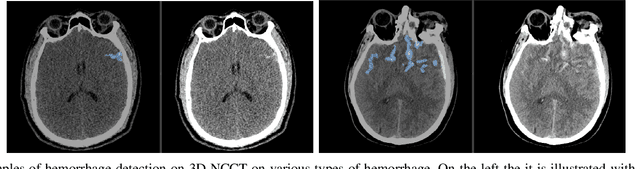
Abstract:Building accurate and robust artificial intelligence systems for medical image assessment requires not only the research and design of advanced deep learning models but also the creation of large and curated sets of annotated training examples. Constructing such datasets, however, is often very costly -- due to the complex nature of annotation tasks and the high level of expertise required for the interpretation of medical images (e.g., expert radiologists). To counter this limitation, we propose a method for self-supervised learning of rich image features based on contrastive learning and online feature clustering. For this purpose we leverage large training datasets of over 100,000,000 medical images of various modalities, including radiography, computed tomography (CT), magnetic resonance (MR) imaging and ultrasonography. We propose to use these features to guide model training in supervised and hybrid self-supervised/supervised regime on various downstream tasks. We highlight a number of advantages of this strategy on challenging image assessment problems in radiography, CT and MR: 1) Significant increase in accuracy compared to the state-of-the-art (e.g., AUC boost of 3-7% for detection of abnormalities from chest radiography scans and hemorrhage detection on brain CT); 2) Acceleration of model convergence during training by up to 85% compared to using no pretraining (e.g., 83% when training a model for detection of brain metastases in MR scans); 3) Increase in robustness to various image augmentations, such as intensity variations, rotations or scaling reflective of data variation seen in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge