Dorin Comaniciu
Revisiting 2D Foundation Models for Scalable 3D Medical Image Classification
Dec 15, 2025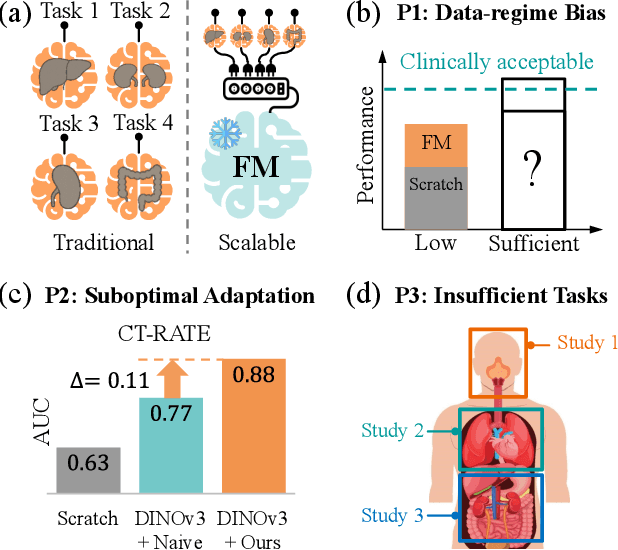
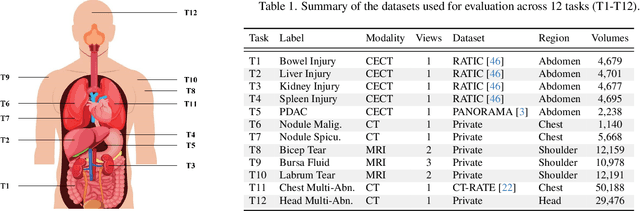
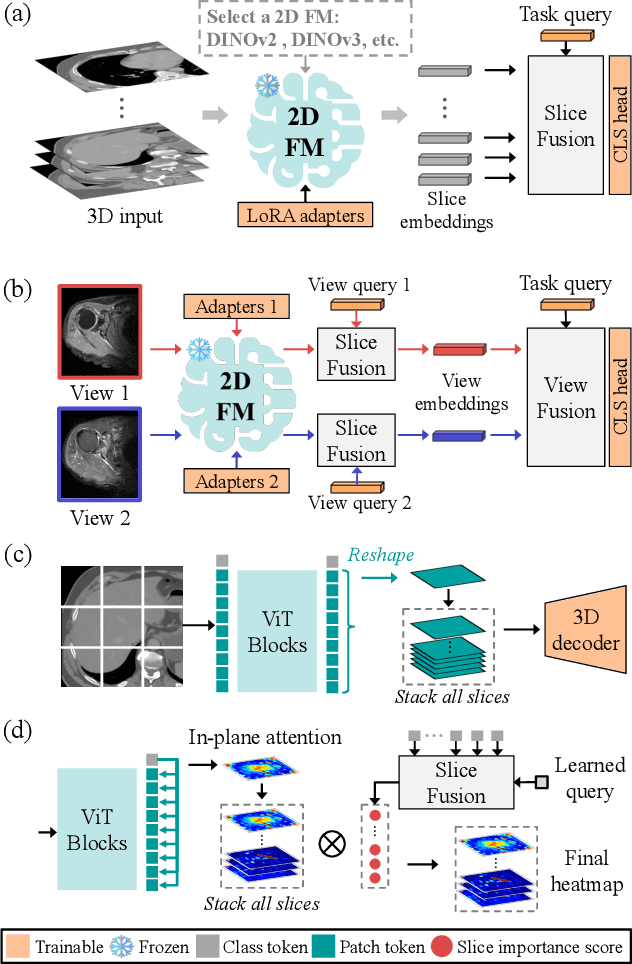
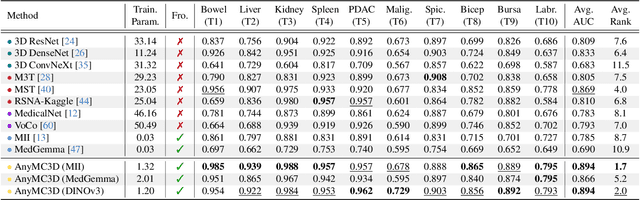
Abstract:3D medical image classification is essential for modern clinical workflows. Medical foundation models (FMs) have emerged as a promising approach for scaling to new tasks, yet current research suffers from three critical pitfalls: data-regime bias, suboptimal adaptation, and insufficient task coverage. In this paper, we address these pitfalls and introduce AnyMC3D, a scalable 3D classifier adapted from 2D FMs. Our method scales efficiently to new tasks by adding only lightweight plugins (about 1M parameters per task) on top of a single frozen backbone. This versatile framework also supports multi-view inputs, auxiliary pixel-level supervision, and interpretable heatmap generation. We establish a comprehensive benchmark of 12 tasks covering diverse pathologies, anatomies, and modalities, and systematically analyze state-of-the-art 3D classification techniques. Our analysis reveals key insights: (1) effective adaptation is essential to unlock FM potential, (2) general-purpose FMs can match medical-specific FMs if properly adapted, and (3) 2D-based methods surpass 3D architectures for 3D classification. For the first time, we demonstrate the feasibility of achieving state-of-the-art performance across diverse applications using a single scalable framework (including 1st place in the VLM3D challenge), eliminating the need for separate task-specific models.
EchoVLM: Measurement-Grounded Multimodal Learning for Echocardiography
Dec 13, 2025Abstract:Echocardiography is the most widely used imaging modality in cardiology, yet its interpretation remains labor-intensive and inherently multimodal, requiring view recognition, quantitative measurements, qualitative assessments, and guideline-based reasoning. While recent vision-language models (VLMs) have achieved broad success in natural images and certain medical domains, their potential in echocardiography has been limited by the lack of large-scale, clinically grounded image-text datasets and the absence of measurement-based reasoning central to echo interpretation. We introduce EchoGround-MIMIC, the first measurement-grounded multimodal echocardiography dataset, comprising 19,065 image-text pairs from 1,572 patients with standardized views, structured measurements, measurement-grounded captions, and guideline-derived disease labels. Building on this resource, we propose EchoVLM, a vision-language model that incorporates two novel pretraining objectives: (i) a view-informed contrastive loss that encodes the view-dependent structure of echocardiographic imaging, and (ii) a negation-aware contrastive loss that distinguishes clinically critical negative from positive findings. Across five types of clinical applications with 36 tasks spanning multimodal disease classification, image-text retrieval, view classification, chamber segmentation, and landmark detection, EchoVLM achieves state-of-the-art performance (86.5% AUC in zero-shot disease classification and 95.1% accuracy in view classification). We demonstrate that clinically grounded multimodal pretraining yields transferable visual representations and establish EchoVLM as a foundation model for end-to-end echocardiography interpretation. We will release EchoGround-MIMIC and the data curation code, enabling reproducibility and further research in multimodal echocardiography interpretation.
Demo: Generative AI helps Radiotherapy Planning with User Preference
Dec 08, 2025Abstract:Radiotherapy planning is a highly complex process that often varies significantly across institutions and individual planners. Most existing deep learning approaches for 3D dose prediction rely on reference plans as ground truth during training, which can inadvertently bias models toward specific planning styles or institutional preferences. In this study, we introduce a novel generative model that predicts 3D dose distributions based solely on user-defined preference flavors. These customizable preferences enable planners to prioritize specific trade-offs between organs-at-risk (OARs) and planning target volumes (PTVs), offering greater flexibility and personalization. Designed for seamless integration with clinical treatment planning systems, our approach assists users in generating high-quality plans efficiently. Comparative evaluations demonstrate that our method can surpasses the Varian RapidPlan model in both adaptability and plan quality in some scenarios.
VIViT: Variable-Input Vision Transformer Framework for 3D MR Image Segmentation
May 13, 2025Abstract:Self-supervised pretrain techniques have been widely used to improve the downstream tasks' performance. However, real-world magnetic resonance (MR) studies usually consist of different sets of contrasts due to different acquisition protocols, which poses challenges for the current deep learning methods on large-scale pretrain and different downstream tasks with different input requirements, since these methods typically require a fixed set of input modalities or, contrasts. To address this challenge, we propose variable-input ViT (VIViT), a transformer-based framework designed for self-supervised pretraining and segmentation finetuning for variable contrasts in each study. With this ability, our approach can maximize the data availability in pretrain, and can transfer the learned knowledge from pretrain to downstream tasks despite variations in input requirements. We validate our method on brain infarct and brain tumor segmentation, where our method outperforms current CNN and ViT-based models with a mean Dice score of 0.624 and 0.883 respectively. These results highlight the efficacy of our design for better adaptability and performance on tasks with real-world heterogeneous MR data.
Multi-Plane Vision Transformer for Hemorrhage Classification Using Axial and Sagittal MRI Data
May 12, 2025Abstract:Identifying brain hemorrhages from magnetic resonance imaging (MRI) is a critical task for healthcare professionals. The diverse nature of MRI acquisitions with varying contrasts and orientation introduce complexity in identifying hemorrhage using neural networks. For acquisitions with varying orientations, traditional methods often involve resampling images to a fixed plane, which can lead to information loss. To address this, we propose a 3D multi-plane vision transformer (MP-ViT) for hemorrhage classification with varying orientation data. It employs two separate transformer encoders for axial and sagittal contrasts, using cross-attention to integrate information across orientations. MP-ViT also includes a modality indication vector to provide missing contrast information to the model. The effectiveness of the proposed model is demonstrated with extensive experiments on real world clinical dataset consists of 10,084 training, 1,289 validation and 1,496 test subjects. MP-ViT achieved substantial improvement in area under the curve (AUC), outperforming the vision transformer (ViT) by 5.5% and CNN-based architectures by 1.8%. These results highlight the potential of MP-ViT in improving performance for hemorrhage detection when different orientation contrasts are needed.
AdaViT: Adaptive Vision Transformer for Flexible Pretrain and Finetune with Variable 3D Medical Image Modalities
Apr 04, 2025



Abstract:Pretrain techniques, whether supervised or self-supervised, are widely used in deep learning to enhance model performance. In real-world clinical scenarios, different sets of magnetic resonance (MR) contrasts are often acquired for different subjects/cases, creating challenges for deep learning models assuming consistent input modalities among all the cases and between pretrain and finetune. Existing methods struggle to maintain performance when there is an input modality/contrast set mismatch with the pretrained model, often resulting in degraded accuracy. We propose an adaptive Vision Transformer (AdaViT) framework capable of handling variable set of input modalities for each case. We utilize a dynamic tokenizer to encode different input image modalities to tokens and take advantage of the characteristics of the transformer to build attention mechanism across variable length of tokens. Through extensive experiments, we demonstrate that this architecture effectively transfers supervised pretrained models to new datasets with different input modality/contrast sets, resulting in superior performance on zero-shot testing, few-shot finetuning, and backward transferring in brain infarct and brain tumor segmentation tasks. Additionally, for self-supervised pretrain, the proposed method is able to maximize the pretrain data and facilitate transferring to diverse downstream tasks with variable sets of input modalities.
Multi-Agent Reinforcement Learning with Long-Term Performance Objectives for Service Workforce Optimization
Mar 03, 2025



Abstract:Workforce optimization plays a crucial role in efficient organizational operations where decision-making may span several different administrative and time scales. For instance, dispatching personnel to immediate service requests while managing talent acquisition with various expertise sets up a highly dynamic optimization problem. Existing work focuses on specific sub-problems such as resource allocation and facility location, which are solved with heuristics like local-search and, more recently, deep reinforcement learning. However, these may not accurately represent real-world scenarios where such sub-problems are not fully independent. Our aim is to fill this gap by creating a simulator that models a unified workforce optimization problem. Specifically, we designed a modular simulator to support the development of reinforcement learning methods for integrated workforce optimization problems. We focus on three interdependent aspects: personnel dispatch, workforce management, and personnel positioning. The simulator provides configurable parameterizations to help explore dynamic scenarios with varying levels of stochasticity and non-stationarity. To facilitate benchmarking and ablation studies, we also include heuristic and RL baselines for the above mentioned aspects.
Self Pre-training with Adaptive Mask Autoencoders for Variable-Contrast 3D Medical Imaging
Jan 15, 2025Abstract:The Masked Autoencoder (MAE) has recently demonstrated effectiveness in pre-training Vision Transformers (ViT) for analyzing natural images. By reconstructing complete images from partially masked inputs, the ViT encoder gathers contextual information to predict the missing regions. This capability to aggregate context is especially important in medical imaging, where anatomical structures are functionally and mechanically linked to surrounding regions. However, current methods do not consider variations in the number of input images, which is typically the case in real-world Magnetic Resonance (MR) studies. To address this limitation, we propose a 3D Adaptive Masked Autoencoders (AMAE) architecture that accommodates a variable number of 3D input contrasts per subject. A magnetic resonance imaging (MRI) dataset of 45,364 subjects was used for pretraining and a subset of 1648 training, 193 validation and 215 test subjects were used for finetuning. The performance demonstrates that self pre-training of this adaptive masked autoencoders can enhance the infarct segmentation performance by 2.8%-3.7% for ViT-based segmentation models.
Towards a vision foundation model for comprehensive assessment of Cardiac MRI
Oct 02, 2024


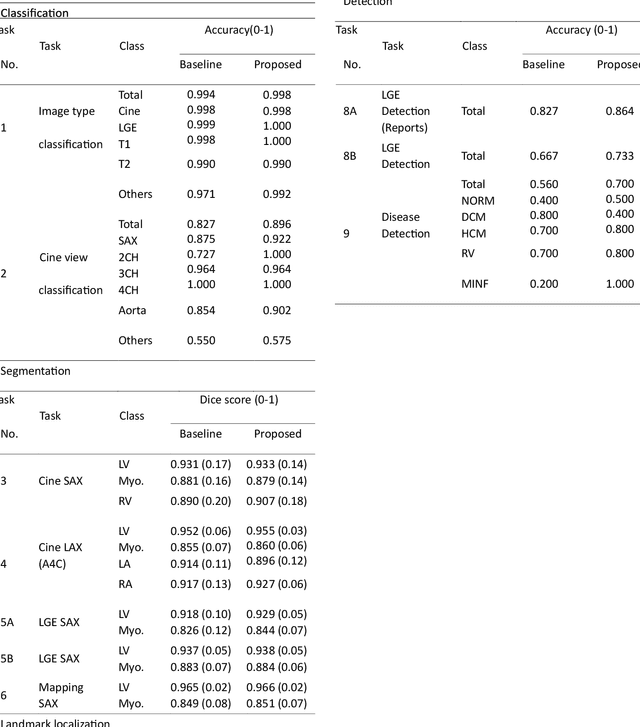
Abstract:Cardiac magnetic resonance imaging (CMR), considered the gold standard for noninvasive cardiac assessment, is a diverse and complex modality requiring a wide variety of image processing tasks for comprehensive assessment of cardiac morphology and function. Advances in deep learning have enabled the development of state-of-the-art (SoTA) models for these tasks. However, model training is challenging due to data and label scarcity, especially in the less common imaging sequences. Moreover, each model is often trained for a specific task, with no connection between related tasks. In this work, we introduce a vision foundation model trained for CMR assessment, that is trained in a self-supervised fashion on 36 million CMR images. We then finetune the model in supervised way for 9 clinical tasks typical to a CMR workflow, across classification, segmentation, landmark localization, and pathology detection. We demonstrate improved accuracy and robustness across all tasks, over a range of available labeled dataset sizes. We also demonstrate improved few-shot learning with fewer labeled samples, a common challenge in medical image analyses. We achieve an out-of-box performance comparable to SoTA for most clinical tasks. The proposed method thus presents a resource-efficient, unified framework for CMR assessment, with the potential to accelerate the development of deep learning-based solutions for image analysis tasks, even with few annotated data available.
Deep Learning-based Unsupervised Domain Adaptation via a Unified Model for Prostate Lesion Detection Using Multisite Bi-parametric MRI Datasets
Aug 08, 2024


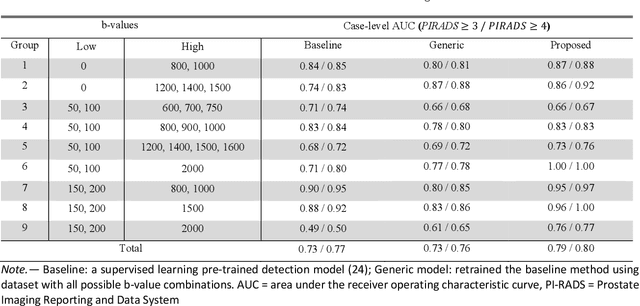
Abstract:Our hypothesis is that UDA using diffusion-weighted images, generated with a unified model, offers a promising and reliable strategy for enhancing the performance of supervised learning models in multi-site prostate lesion detection, especially when various b-values are present. This retrospective study included data from 5,150 patients (14,191 samples) collected across nine different imaging centers. A novel UDA method using a unified generative model was developed for multi-site PCa detection. This method translates diffusion-weighted imaging (DWI) acquisitions, including apparent diffusion coefficient (ADC) and individual DW images acquired using various b-values, to align with the style of images acquired using b-values recommended by Prostate Imaging Reporting and Data System (PI-RADS) guidelines. The generated ADC and DW images replace the original images for PCa detection. An independent set of 1,692 test cases (2,393 samples) was used for evaluation. The area under the receiver operating characteristic curve (AUC) was used as the primary metric, and statistical analysis was performed via bootstrapping. For all test cases, the AUC values for baseline SL and UDA methods were 0.73 and 0.79 (p<.001), respectively, for PI-RADS>=3, and 0.77 and 0.80 (p<.001) for PI-RADS>=4 PCa lesions. In the 361 test cases under the most unfavorable image acquisition setting, the AUC values for baseline SL and UDA were 0.49 and 0.76 (p<.001) for PI-RADS>=3, and 0.50 and 0.77 (p<.001) for PI-RADS>=4 PCa lesions. The results indicate the proposed UDA with generated images improved the performance of SL methods in multi-site PCa lesion detection across datasets with various b values, especially for images acquired with significant deviations from the PI-RADS recommended DWI protocol (e.g. with an extremely high b-value).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge