Teodora Chitiboi
Deep learning-based segmentation of T1 and T2 cardiac MRI maps for automated disease detection
Jul 01, 2025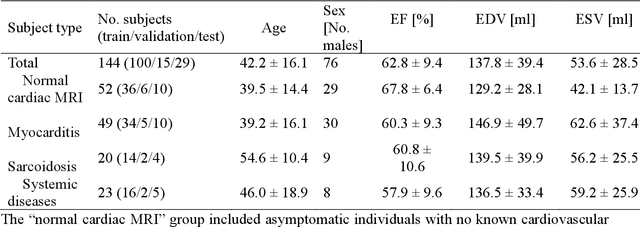
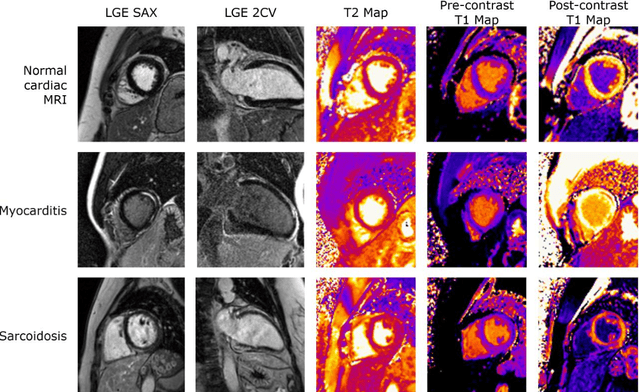
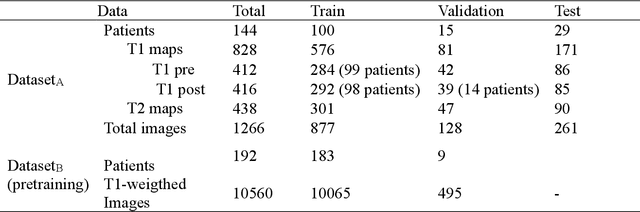
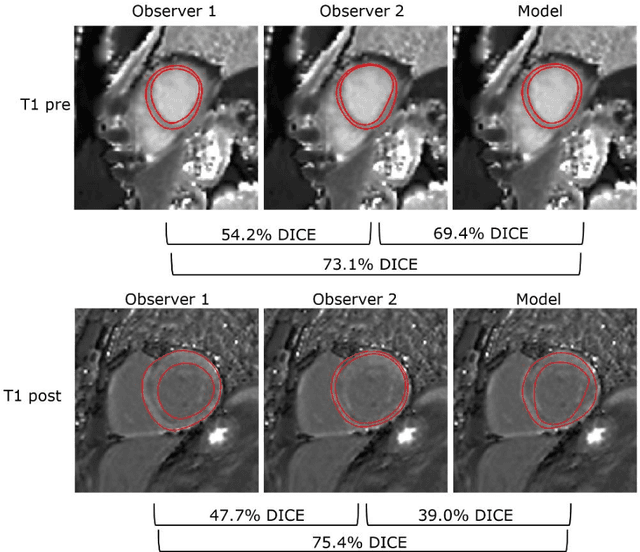
Abstract:Objectives Parametric tissue mapping enables quantitative cardiac tissue characterization but is limited by inter-observer variability during manual delineation. Traditional approaches relying on average relaxation values and single cutoffs may oversimplify myocardial complexity. This study evaluates whether deep learning (DL) can achieve segmentation accuracy comparable to inter-observer variability, explores the utility of statistical features beyond mean T1/T2 values, and assesses whether machine learning (ML) combining multiple features enhances disease detection. Materials & Methods T1 and T2 maps were manually segmented. The test subset was independently annotated by two observers, and inter-observer variability was assessed. A DL model was trained to segment left ventricle blood pool and myocardium. Average (A), lower quartile (LQ), median (M), and upper quartile (UQ) were computed for the myocardial pixels and employed in classification by applying cutoffs or in ML. Dice similarity coefficient (DICE) and mean absolute percentage error evaluated segmentation performance. Bland-Altman plots assessed inter-user and model-observer agreement. Receiver operating characteristic analysis determined optimal cutoffs. Pearson correlation compared features from model and manual segmentations. F1-score, precision, and recall evaluated classification performance. Wilcoxon test assessed differences between classification methods, with p < 0.05 considered statistically significant. Results 144 subjects were split into training (100), validation (15) and evaluation (29) subsets. Segmentation model achieved a DICE of 85.4%, surpassing inter-observer agreement. Random forest applied to all features increased F1-score (92.7%, p < 0.001). Conclusion DL facilitates segmentation of T1/ T2 maps. Combining multiple features with ML improves disease detection.
Towards a vision foundation model for comprehensive assessment of Cardiac MRI
Oct 02, 2024


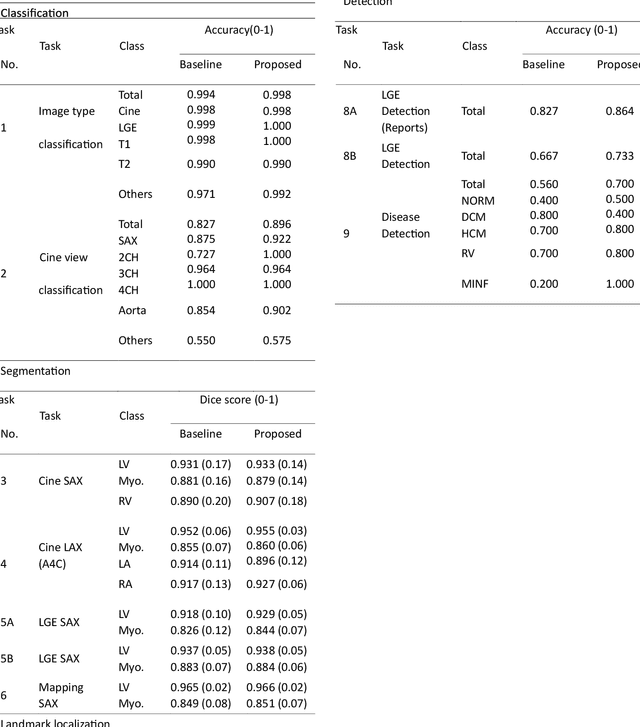
Abstract:Cardiac magnetic resonance imaging (CMR), considered the gold standard for noninvasive cardiac assessment, is a diverse and complex modality requiring a wide variety of image processing tasks for comprehensive assessment of cardiac morphology and function. Advances in deep learning have enabled the development of state-of-the-art (SoTA) models for these tasks. However, model training is challenging due to data and label scarcity, especially in the less common imaging sequences. Moreover, each model is often trained for a specific task, with no connection between related tasks. In this work, we introduce a vision foundation model trained for CMR assessment, that is trained in a self-supervised fashion on 36 million CMR images. We then finetune the model in supervised way for 9 clinical tasks typical to a CMR workflow, across classification, segmentation, landmark localization, and pathology detection. We demonstrate improved accuracy and robustness across all tasks, over a range of available labeled dataset sizes. We also demonstrate improved few-shot learning with fewer labeled samples, a common challenge in medical image analyses. We achieve an out-of-box performance comparable to SoTA for most clinical tasks. The proposed method thus presents a resource-efficient, unified framework for CMR assessment, with the potential to accelerate the development of deep learning-based solutions for image analysis tasks, even with few annotated data available.
Automated Cardiac Resting Phase Detection Targeted on the Right Coronary Artery
Sep 06, 2021



Abstract:Purpose: Static cardiac imaging such as late gadolinium enhancement, mapping, or 3-D coronary angiography require prior information, e.g., the phase during a cardiac cycle with least motion, called resting phase (RP). The purpose of this work is to propose a fully automated framework that allows the detection of the right coronary artery (RCA) RP within CINE series. Methods: The proposed prototype system consists of three main steps. First, the localization of the regions of interest (ROI) is performed. Second, as CINE series are time-resolved, the cropped ROI series over all time points are taken for tracking motions quantitatively. Third, the output motion values are used to classify RPs. In this work, we focused on the detection of the area with the outer edge of the cross-section of the RCA as our target. The proposed framework was evaluated on 102 clinically acquired dataset at 1.5T and 3T. The automatically classified RPs were compared with the ground truth RPs annotated manually by a medical expert for testing the robustness and feasibility of the framework. Results: The predicted RCA RPs showed high agreement with the experts annotated RPs with 92.7% accuracy, 90.5% sensitivity and 95.0% specificity for the unseen study dataset. The mean absolute difference of the start and end RP was 13.6 ${\pm}$ 18.6 ms for the validation study dataset (n=102). Conclusion: In this work, automated RP detection has been introduced by the proposed framework and demonstrated feasibility, robustness, and applicability for diverse static imaging acquisitions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge