Puneet Sharma
Using Multi-Instance Learning to Identify Unique Polyps in Colon Capsule Endoscopy Images
Jan 21, 2026Abstract:Identifying unique polyps in colon capsule endoscopy (CCE) images is a critical yet challenging task for medical personnel due to the large volume of images, the cognitive load it creates for clinicians, and the ambiguity in labeling specific frames. This paper formulates this problem as a multi-instance learning (MIL) task, where a query polyp image is compared with a target bag of images to determine uniqueness. We employ a multi-instance verification (MIV) framework that incorporates attention mechanisms, such as variance-excited multi-head attention (VEMA) and distance-based attention (DBA), to enhance the model's ability to extract meaningful representations. Additionally, we investigate the impact of self-supervised learning using SimCLR to generate robust embeddings. Experimental results on a dataset of 1912 polyps from 754 patients demonstrate that attention mechanisms significantly improve performance, with DBA L1 achieving the highest test accuracy of 86.26\% and a test AUC of 0.928 using a ConvNeXt backbone with SimCLR pretraining. This study underscores the potential of MIL and self-supervised learning in advancing automated analysis of Colon Capsule Endoscopy images, with implications for broader medical imaging applications.
Using deep learning for predicting cleansing quality of colon capsule endoscopy images
Jan 19, 2026Abstract:In this study, we explore the application of deep learning techniques for predicting cleansing quality in colon capsule endoscopy (CCE) images. Using a dataset of 500 images labeled by 14 clinicians on the Leighton-Rex scale (Poor, Fair, Good, and Excellent), a ResNet-18 model was trained for classification, leveraging stratified K-fold cross-validation to ensure robust performance. To optimize the model, structured pruning techniques were applied iteratively, achieving significant sparsity while maintaining high accuracy. Explainability of the pruned model was evaluated using Grad-CAM, Grad-CAM++, Eigen-CAM, Ablation-CAM, and Random-CAM, with the ROAD method employed for consistent evaluation. Our results indicate that for a pruned model, we can achieve a cross-validation accuracy of 88% with 79% sparsity, demonstrating the effectiveness of pruning in improving efficiency from 84% without compromising performance. We also highlight the challenges of evaluating cleansing quality of CCE images, emphasize the importance of explainability in clinical applications, and discuss the challenges associated with using the ROAD method for our task. Finally, we employ a variant of adaptive temperature scaling to calibrate the pruned models for an external dataset.
EchoVLM: Measurement-Grounded Multimodal Learning for Echocardiography
Dec 13, 2025Abstract:Echocardiography is the most widely used imaging modality in cardiology, yet its interpretation remains labor-intensive and inherently multimodal, requiring view recognition, quantitative measurements, qualitative assessments, and guideline-based reasoning. While recent vision-language models (VLMs) have achieved broad success in natural images and certain medical domains, their potential in echocardiography has been limited by the lack of large-scale, clinically grounded image-text datasets and the absence of measurement-based reasoning central to echo interpretation. We introduce EchoGround-MIMIC, the first measurement-grounded multimodal echocardiography dataset, comprising 19,065 image-text pairs from 1,572 patients with standardized views, structured measurements, measurement-grounded captions, and guideline-derived disease labels. Building on this resource, we propose EchoVLM, a vision-language model that incorporates two novel pretraining objectives: (i) a view-informed contrastive loss that encodes the view-dependent structure of echocardiographic imaging, and (ii) a negation-aware contrastive loss that distinguishes clinically critical negative from positive findings. Across five types of clinical applications with 36 tasks spanning multimodal disease classification, image-text retrieval, view classification, chamber segmentation, and landmark detection, EchoVLM achieves state-of-the-art performance (86.5% AUC in zero-shot disease classification and 95.1% accuracy in view classification). We demonstrate that clinically grounded multimodal pretraining yields transferable visual representations and establish EchoVLM as a foundation model for end-to-end echocardiography interpretation. We will release EchoGround-MIMIC and the data curation code, enabling reproducibility and further research in multimodal echocardiography interpretation.
Towards Easy and Realistic Network Infrastructure Testing for Large-scale Machine Learning
Apr 29, 2025


Abstract:This paper lays the foundation for Genie, a testing framework that captures the impact of real hardware network behavior on ML workload performance, without requiring expensive GPUs. Genie uses CPU-initiated traffic over a hardware testbed to emulate GPU to GPU communication, and adapts the ASTRA-sim simulator to model interaction between the network and the ML workload.
Fake It Till You Make It: Using Synthetic Data and Domain Knowledge for Improved Text-Based Learning for LGE Detection
Feb 18, 2025



Abstract:Detection of hyperenhancement from cardiac LGE MRI images is a complex task requiring significant clinical expertise. Although deep learning-based models have shown promising results for the task, they require large amounts of data with fine-grained annotations. Clinical reports generated for cardiac MR studies contain rich, clinically relevant information, including the location, extent and etiology of any scars present. Although recently developed CLIP-based training enables pretraining models with image-text pairs, it requires large amounts of data and further finetuning strategies on downstream tasks. In this study, we use various strategies rooted in domain knowledge to train a model for LGE detection solely using text from clinical reports, on a relatively small clinical cohort of 965 patients. We improve performance through the use of synthetic data augmentation, by systematically creating scar images and associated text. In addition, we standardize the orientation of the images in an anatomy-informed way to enable better alignment of spatial and text features. We also use a captioning loss to enable fine-grained supervision and explore the effect of pretraining of the vision encoder on performance. Finally, ablation studies are carried out to elucidate the contributions of each design component to the overall performance of the model.
EchoApex: A General-Purpose Vision Foundation Model for Echocardiography
Oct 14, 2024



Abstract:Quantitative evaluation of echocardiography is essential for precise assessment of cardiac condition, monitoring disease progression, and guiding treatment decisions. The diverse nature of echo images, including variations in probe types, manufacturers, and pathologies, poses challenges for developing artificial intelligent models that can generalize across different clinical practice. We introduce EchoApex, the first general-purpose vision foundation model echocardiography with applications on a variety of clinical practice. Leveraging self-supervised learning, EchoApex is pretrained on over 20 million echo images from 11 clinical centres. By incorporating task-specific decoders and adapter modules, we demonstrate the effectiveness of EchoApex on 4 different kind of clinical applications with 28 sub-tasks, including view classification, interactive structure segmentation, left ventricle hypertrophy detection and automated ejection fraction estimation from view sequences. Compared to state-of-the-art task-specific models, EchoApex attains improved performance with a unified image encoding architecture, demonstrating the benefits of model pretraining at scale with in-domain data. Furthermore, EchoApex illustrates the potential for developing a general-purpose vision foundation model tailored specifically for echocardiography, capable of addressing a diverse range of clinical applications with high efficiency and efficacy.
Towards a vision foundation model for comprehensive assessment of Cardiac MRI
Oct 02, 2024


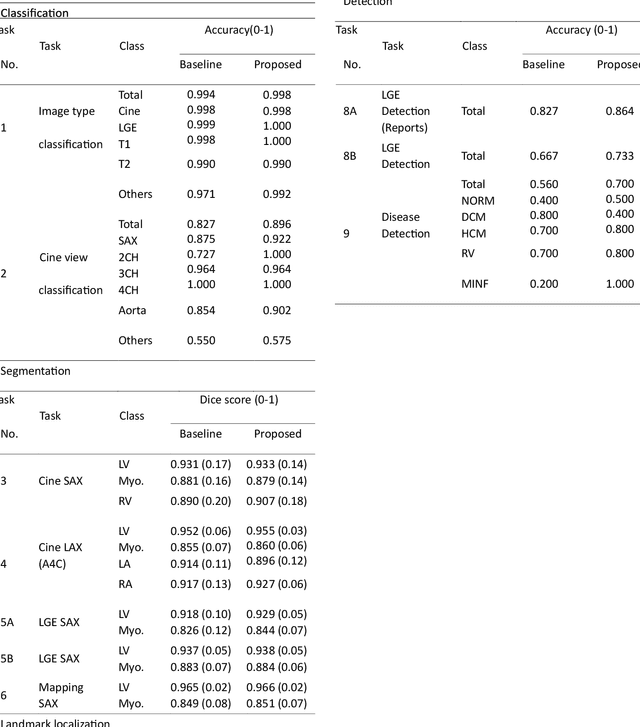
Abstract:Cardiac magnetic resonance imaging (CMR), considered the gold standard for noninvasive cardiac assessment, is a diverse and complex modality requiring a wide variety of image processing tasks for comprehensive assessment of cardiac morphology and function. Advances in deep learning have enabled the development of state-of-the-art (SoTA) models for these tasks. However, model training is challenging due to data and label scarcity, especially in the less common imaging sequences. Moreover, each model is often trained for a specific task, with no connection between related tasks. In this work, we introduce a vision foundation model trained for CMR assessment, that is trained in a self-supervised fashion on 36 million CMR images. We then finetune the model in supervised way for 9 clinical tasks typical to a CMR workflow, across classification, segmentation, landmark localization, and pathology detection. We demonstrate improved accuracy and robustness across all tasks, over a range of available labeled dataset sizes. We also demonstrate improved few-shot learning with fewer labeled samples, a common challenge in medical image analyses. We achieve an out-of-box performance comparable to SoTA for most clinical tasks. The proposed method thus presents a resource-efficient, unified framework for CMR assessment, with the potential to accelerate the development of deep learning-based solutions for image analysis tasks, even with few annotated data available.
AI-driven View Guidance System in Intra-cardiac Echocardiography Imaging
Sep 26, 2024



Abstract:Intra-cardiac Echocardiography (ICE) is a crucial imaging modality used in electrophysiology (EP) and structural heart disease (SHD) interventions, providing real-time, high-resolution views from within the heart. Despite its advantages, effective manipulation of the ICE catheter requires significant expertise, which can lead to inconsistent outcomes, particularly among less experienced operators. To address this challenge, we propose an AI-driven closed-loop view guidance system with human-in-the-loop feedback, designed to assist users in navigating ICE imaging without requiring specialized knowledge. Our method models the relative position and orientation vectors between arbitrary views and clinically defined ICE views in a spatial coordinate system, guiding users on how to manipulate the ICE catheter to transition from the current view to the desired view over time. Operating in a closed-loop configuration, the system continuously predicts and updates the necessary catheter manipulations, ensuring seamless integration into existing clinical workflows. The effectiveness of the proposed system is demonstrated through a simulation-based evaluation, achieving an 89% success rate with the 6532 test dataset, highlighting its potential to improve the accuracy and efficiency of ICE imaging procedures.
DCSM 2.0: Deep Conditional Shape Models for Data Efficient Segmentation
Jun 28, 2024


Abstract:Segmentation is often the first step in many medical image analyses workflows. Deep learning approaches, while giving state-of-the-art accuracies, are data intensive and do not scale well to low data regimes. We introduce Deep Conditional Shape Models 2.0, which uses an edge detector, along with an implicit shape function conditioned on edge maps, to leverage cross-modality shape information. The shape function is trained exclusively on a source domain (contrasted CT) and applied to the target domain of interest (3D echocardiography). We demonstrate data efficiency in the target domain by varying the amounts of training data used in the edge detection stage. We observe that DCSM 2.0 outperforms the baseline at all data levels in terms of Hausdorff distances, and while using 50% or less of the training data in terms of average mesh distance, and at 10% or less of the data with the dice coefficient. The method scales well to low data regimes, with gains of up to 5% in dice coefficient, 2.58 mm in average surface distance and 21.02 mm in Hausdorff distance when using just 2% (22 volumes) of the training data.
Goal-conditioned reinforcement learning for ultrasound navigation guidance
May 02, 2024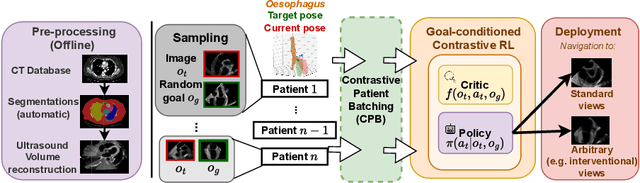
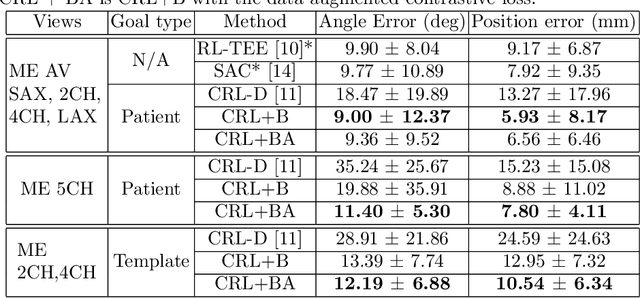
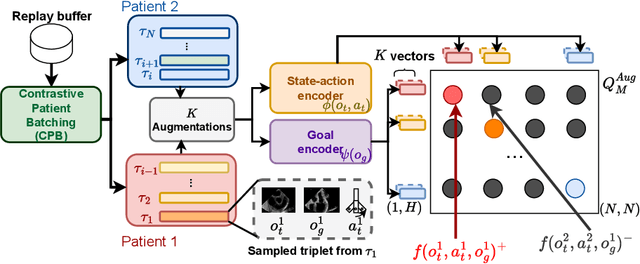
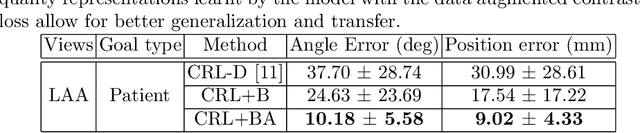
Abstract:Transesophageal echocardiography (TEE) plays a pivotal role in cardiology for diagnostic and interventional procedures. However, using it effectively requires extensive training due to the intricate nature of image acquisition and interpretation. To enhance the efficiency of novice sonographers and reduce variability in scan acquisitions, we propose a novel ultrasound (US) navigation assistance method based on contrastive learning as goal-conditioned reinforcement learning (GCRL). We augment the previous framework using a novel contrastive patient batching method (CPB) and a data-augmented contrastive loss, both of which we demonstrate are essential to ensure generalization to anatomical variations across patients. The proposed framework enables navigation to both standard diagnostic as well as intricate interventional views with a single model. Our method was developed with a large dataset of 789 patients and obtained an average error of 6.56 mm in position and 9.36 degrees in angle on a testing dataset of 140 patients, which is competitive or superior to models trained on individual views. Furthermore, we quantitatively validate our method's ability to navigate to interventional views such as the Left Atrial Appendage (LAA) view used in LAA closure. Our approach holds promise in providing valuable guidance during transesophageal ultrasound examinations, contributing to the advancement of skill acquisition for cardiac ultrasound practitioners.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge