Alistair Young
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Implicit Neural Representations of Intramyocardial Motion and Strain
Sep 10, 2025Abstract:Automatic quantification of intramyocardial motion and strain from tagging MRI remains an important but challenging task. We propose a method using implicit neural representations (INRs), conditioned on learned latent codes, to predict continuous left ventricular (LV) displacement -- without requiring inference-time optimisation. Evaluated on 452 UK Biobank test cases, our method achieved the best tracking accuracy (2.14 mm RMSE) and the lowest combined error in global circumferential (2.86%) and radial (6.42%) strain compared to three deep learning baselines. In addition, our method is $\sim$380$\times$ faster than the most accurate baseline. These results highlight the suitability of INR-based models for accurate and scalable analysis of myocardial strain in large CMR datasets.
Cardiac Digital Twins at Scale from MRI: Open Tools and Representative Models from ~55000 UK Biobank Participants
May 27, 2025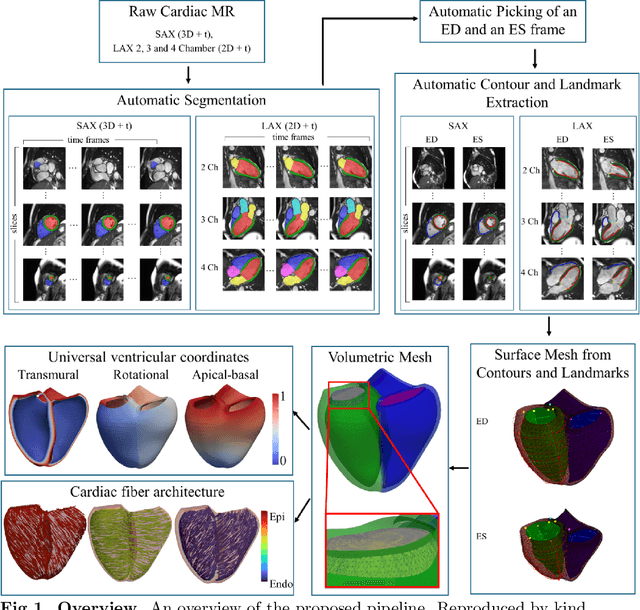
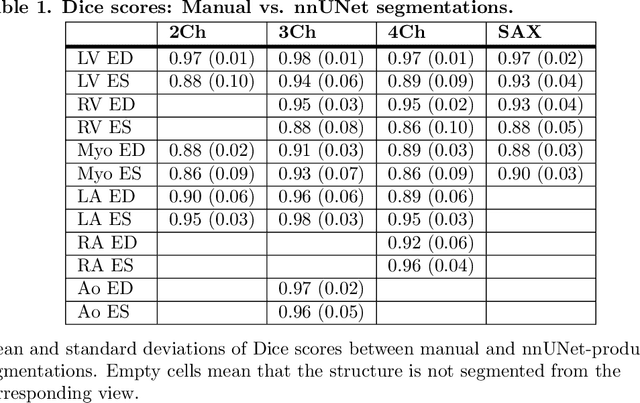
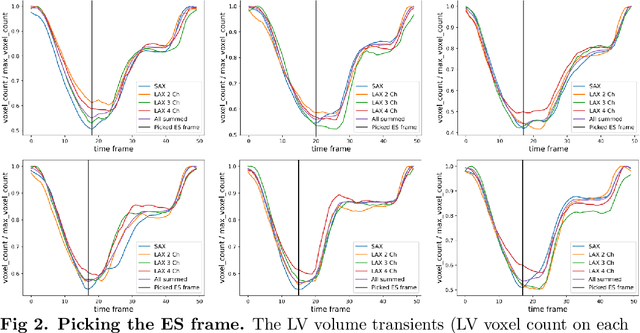
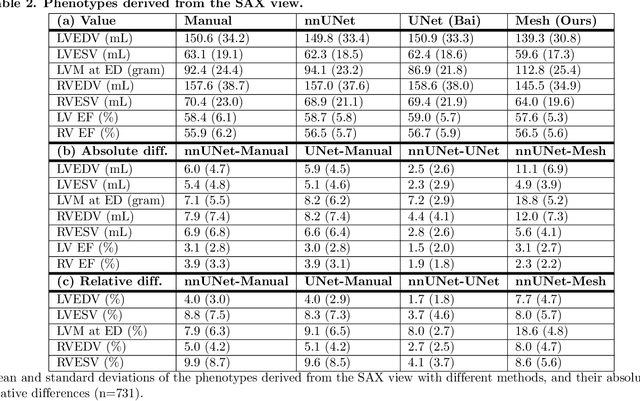
Abstract:A cardiac digital twin is a virtual replica of a patient's heart for screening, diagnosis, prognosis, risk assessment, and treatment planning of cardiovascular diseases. This requires an anatomically accurate patient-specific 3D structural representation of the heart, suitable for electro-mechanical simulations or study of disease mechanisms. However, generation of cardiac digital twins at scale is demanding and there are no public repositories of models across demographic groups. We describe an automatic open-source pipeline for creating patient-specific left and right ventricular meshes from cardiovascular magnetic resonance images, its application to a large cohort of ~55000 participants from UK Biobank, and the construction of the most comprehensive cohort of adult heart models to date, comprising 1423 representative meshes across sex (male, female), body mass index (range: 16 - 42 kg/m$^2$) and age (range: 49 - 80 years). Our code is available at https://github.com/cdttk/biv-volumetric-meshing/tree/plos2025 , and pre-trained networks, representative volumetric meshes with fibers and UVCs will be made available soon.
A Computational Pipeline for Advanced Analysis of 4D Flow MRI in the Left Atrium
May 14, 2025Abstract:The left atrium (LA) plays a pivotal role in modulating left ventricular filling, but our comprehension of its hemodynamics is significantly limited by the constraints of conventional ultrasound analysis. 4D flow magnetic resonance imaging (4D Flow MRI) holds promise for enhancing our understanding of atrial hemodynamics. However, the low velocities within the LA and the limited spatial resolution of 4D Flow MRI make analyzing this chamber challenging. Furthermore, the absence of dedicated computational frameworks, combined with diverse acquisition protocols and vendors, complicates gathering large cohorts for studying the prognostic value of hemodynamic parameters provided by 4D Flow MRI. In this study, we introduce the first open-source computational framework tailored for the analysis of 4D Flow MRI in the LA, enabling comprehensive qualitative and quantitative analysis of advanced hemodynamic parameters. Our framework proves robust to data from different centers of varying quality, producing high-accuracy automated segmentations (Dice $>$ 0.9 and Hausdorff 95 $<$ 3 mm), even with limited training data. Additionally, we conducted the first comprehensive assessment of energy, vorticity, and pressure parameters in the LA across a spectrum of disorders to investigate their potential as prognostic biomarkers.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
MorphiNet: A Graph Subdivision Network for Adaptive Bi-ventricle Surface Reconstruction
Dec 14, 2024



Abstract:Cardiac Magnetic Resonance (CMR) imaging is widely used for heart modelling and digital twin computational analysis due to its ability to visualize soft tissues and capture dynamic functions. However, the anisotropic nature of CMR images, characterized by large inter-slice distances and misalignments from cardiac motion, poses significant challenges to accurate model reconstruction. These limitations result in data loss and measurement inaccuracies, hindering the capture of detailed anatomical structures. This study introduces MorphiNet, a novel network that enhances heart model reconstruction by leveraging high-resolution Computer Tomography (CT) images, unpaired with CMR images, to learn heart anatomy. MorphiNet encodes anatomical structures as gradient fields, transforming template meshes into patient-specific geometries. A multi-layer graph subdivision network refines these geometries while maintaining dense point correspondence. The proposed method achieves high anatomy fidelity, demonstrating approximately 40% higher Dice scores, half the Hausdorff distance, and around 3 mm average surface error compared to state-of-the-art methods. MorphiNet delivers superior results with greater inference efficiency. This approach represents a significant advancement in addressing the challenges of CMR-based heart model reconstruction, potentially improving digital twin computational analyses of cardiac structure and functions.
CMRxRecon2024: A Multi-Modality, Multi-View K-Space Dataset Boosting Universal Machine Learning for Accelerated Cardiac MRI
Jun 27, 2024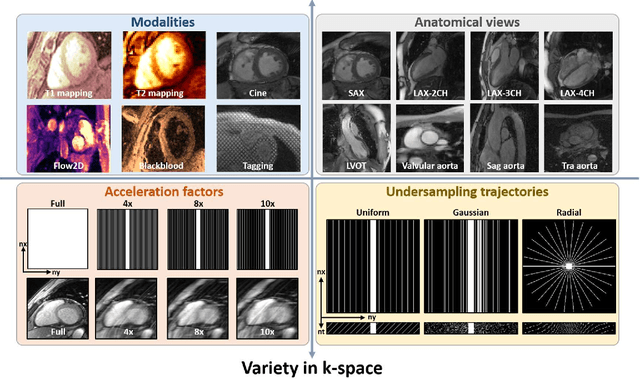
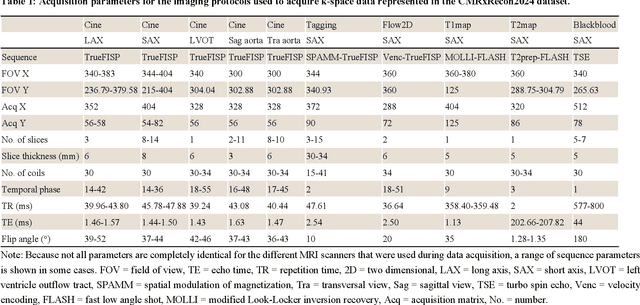
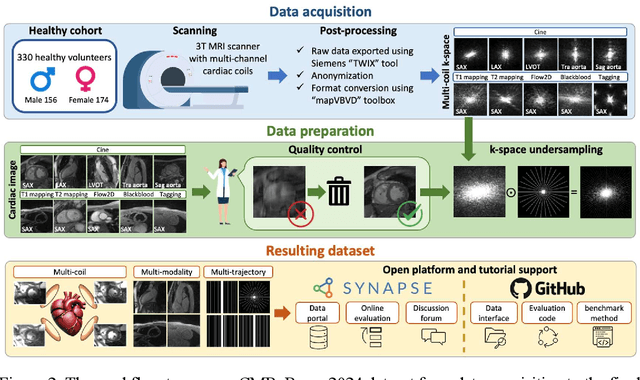
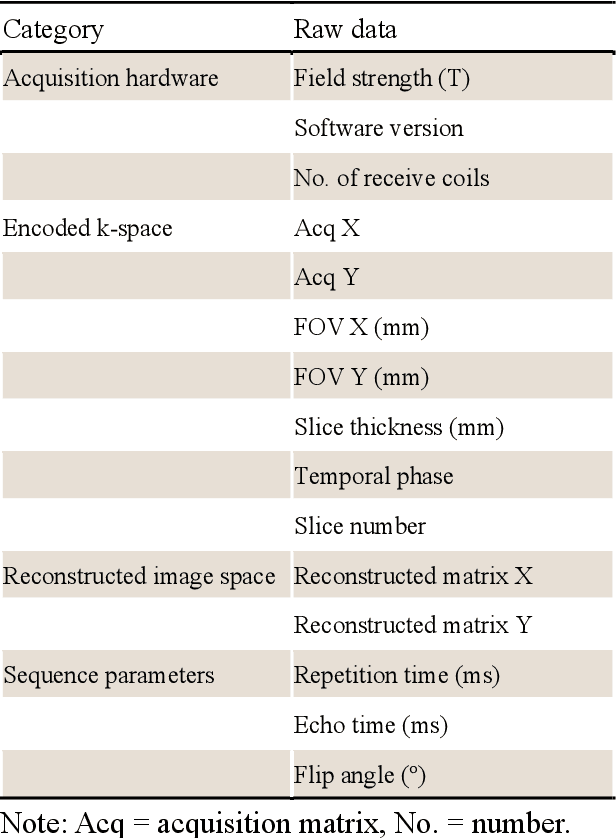
Abstract:Cardiac magnetic resonance imaging (MRI) has emerged as a clinically gold-standard technique for diagnosing cardiac diseases, thanks to its ability to provide diverse information with multiple modalities and anatomical views. Accelerated cardiac MRI is highly expected to achieve time-efficient and patient-friendly imaging, and then advanced image reconstruction approaches are required to recover high-quality, clinically interpretable images from undersampled measurements. However, the lack of publicly available cardiac MRI k-space dataset in terms of both quantity and diversity has severely hindered substantial technological progress, particularly for data-driven artificial intelligence. Here, we provide a standardized, diverse, and high-quality CMRxRecon2024 dataset to facilitate the technical development, fair evaluation, and clinical transfer of cardiac MRI reconstruction approaches, towards promoting the universal frameworks that enable fast and robust reconstructions across different cardiac MRI protocols in clinical practice. To the best of our knowledge, the CMRxRecon2024 dataset is the largest and most diverse publicly available cardiac k-space dataset. It is acquired from 330 healthy volunteers, covering commonly used modalities, anatomical views, and acquisition trajectories in clinical cardiac MRI workflows. Besides, an open platform with tutorials, benchmarks, and data processing tools is provided to facilitate data usage, advanced method development, and fair performance evaluation.
Goal-conditioned reinforcement learning for ultrasound navigation guidance
May 02, 2024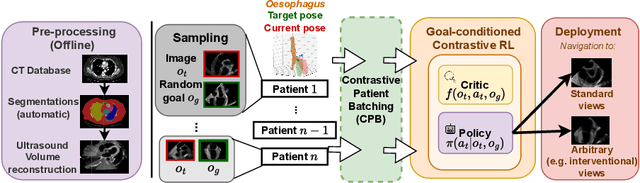
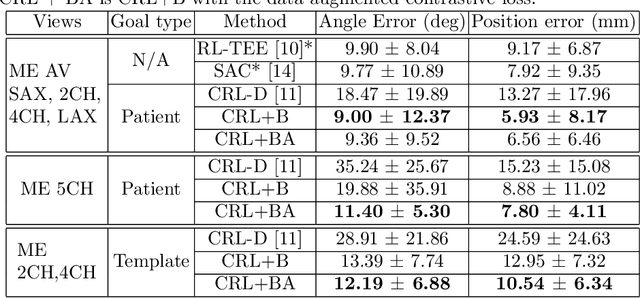
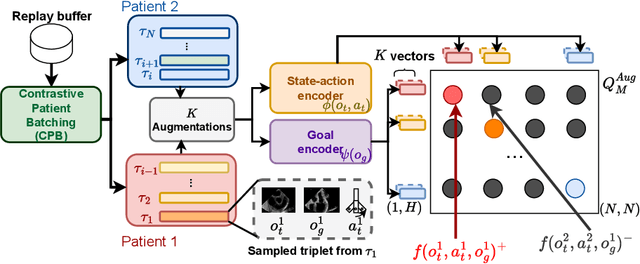
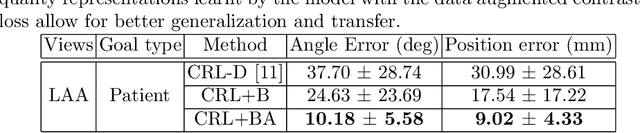
Abstract:Transesophageal echocardiography (TEE) plays a pivotal role in cardiology for diagnostic and interventional procedures. However, using it effectively requires extensive training due to the intricate nature of image acquisition and interpretation. To enhance the efficiency of novice sonographers and reduce variability in scan acquisitions, we propose a novel ultrasound (US) navigation assistance method based on contrastive learning as goal-conditioned reinforcement learning (GCRL). We augment the previous framework using a novel contrastive patient batching method (CPB) and a data-augmented contrastive loss, both of which we demonstrate are essential to ensure generalization to anatomical variations across patients. The proposed framework enables navigation to both standard diagnostic as well as intricate interventional views with a single model. Our method was developed with a large dataset of 789 patients and obtained an average error of 6.56 mm in position and 9.36 degrees in angle on a testing dataset of 140 patients, which is competitive or superior to models trained on individual views. Furthermore, we quantitatively validate our method's ability to navigate to interventional views such as the Left Atrial Appendage (LAA) view used in LAA closure. Our approach holds promise in providing valuable guidance during transesophageal ultrasound examinations, contributing to the advancement of skill acquisition for cardiac ultrasound practitioners.
Cardiac ultrasound simulation for autonomous ultrasound navigation
Feb 09, 2024Abstract:Ultrasound is well-established as an imaging modality for diagnostic and interventional purposes. However, the image quality varies with operator skills as acquiring and interpreting ultrasound images requires extensive training due to the imaging artefacts, the range of acquisition parameters and the variability of patient anatomies. Automating the image acquisition task could improve acquisition reproducibility and quality but training such an algorithm requires large amounts of navigation data, not saved in routine examinations. Thus, we propose a method to generate large amounts of ultrasound images from other modalities and from arbitrary positions, such that this pipeline can later be used by learning algorithms for navigation. We present a novel simulation pipeline which uses segmentations from other modalities, an optimized volumetric data representation and GPU-accelerated Monte Carlo path tracing to generate view-dependent and patient-specific ultrasound images. We extensively validate the correctness of our pipeline with a phantom experiment, where structures' sizes, contrast and speckle noise properties are assessed. Furthermore, we demonstrate its usability to train neural networks for navigation in an echocardiography view classification experiment by generating synthetic images from more than 1000 patients. Networks pre-trained with our simulations achieve significantly superior performance in settings where large real datasets are not available, especially for under-represented classes. The proposed approach allows for fast and accurate patient-specific ultrasound image generation, and its usability for training networks for navigation-related tasks is demonstrated.
The Impact of Domain Shift on Left and Right Ventricle Segmentation in Short Axis Cardiac MR Images
Sep 22, 2021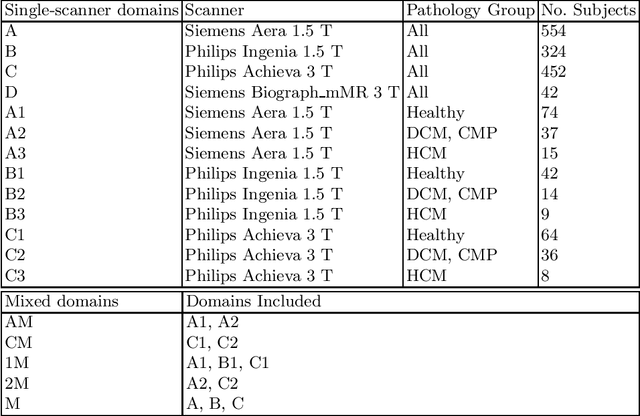
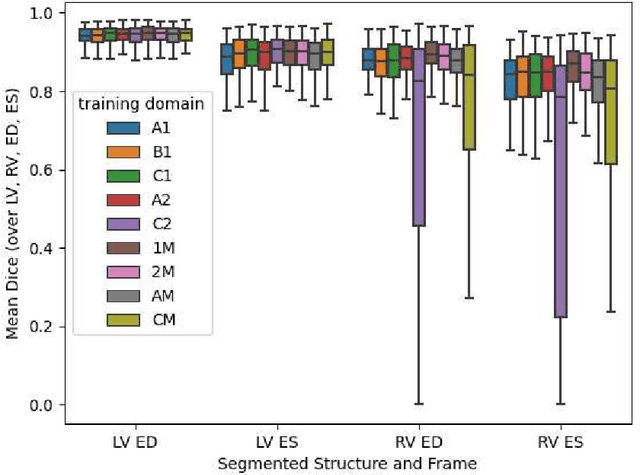
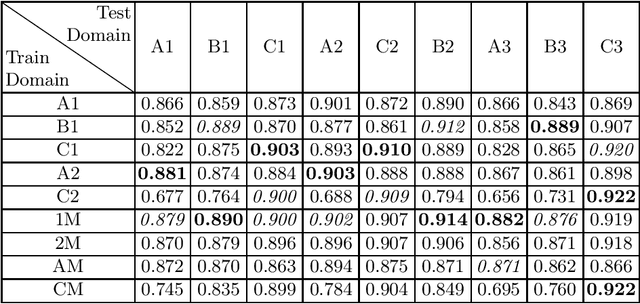

Abstract:Domain shift refers to the difference in the data distribution of two datasets, normally between the training set and the test set for machine learning algorithms. Domain shift is a serious problem for generalization of machine learning models and it is well-established that a domain shift between the training and test sets may cause a drastic drop in the model's performance. In medical imaging, there can be many sources of domain shift such as different scanners or scan protocols, different pathologies in the patient population, anatomical differences in the patient population (e.g. men vs women) etc. Therefore, in order to train models that have good generalization performance, it is important to be aware of the domain shift problem, its potential causes and to devise ways to address it. In this paper, we study the effect of domain shift on left and right ventricle blood pool segmentation in short axis cardiac MR images. Our dataset contains short axis images from 4 different MR scanners and 3 different pathology groups. The training is performed with nnUNet. The results show that scanner differences cause a greater drop in performance compared to changing the pathology group, and that the impact of domain shift is greater on right ventricle segmentation compared to left ventricle segmentation. Increasing the number of training subjects increased cross-scanner performance more than in-scanner performance at small training set sizes, but this difference in improvement decreased with larger training set sizes. Training models using data from multiple scanners improved cross-domain performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge