Esther Puyol-Anton
An investigation into the causes of race bias in AI-based cine CMR segmentation
Aug 05, 2024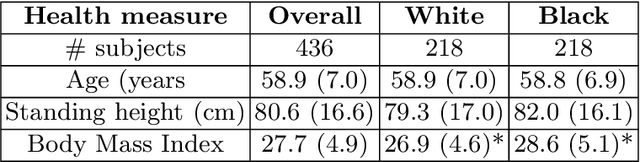
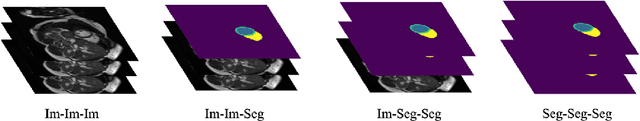
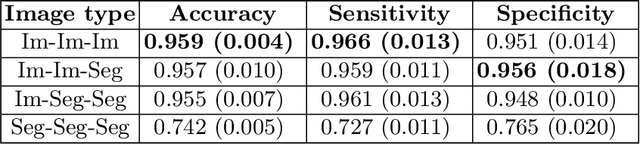
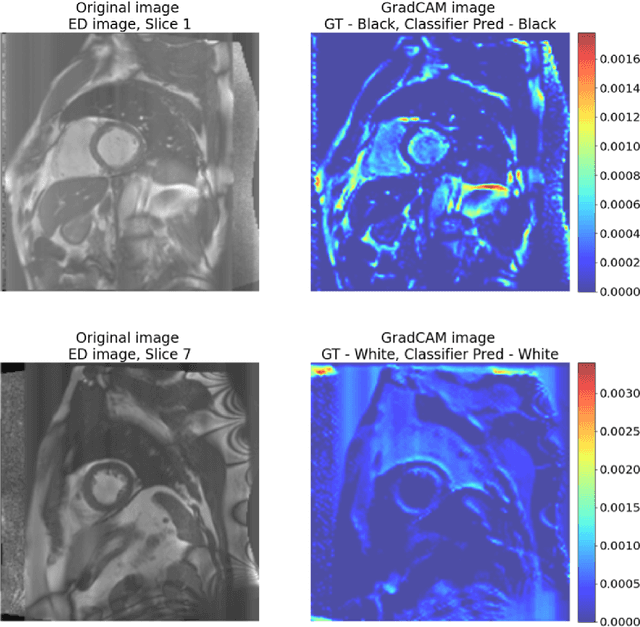
Abstract:Artificial intelligence (AI) methods are being used increasingly for the automated segmentation of cine cardiac magnetic resonance (CMR) imaging. However, these methods have been shown to be subject to race bias, i.e. they exhibit different levels of performance for different races depending on the (im)balance of the data used to train the AI model. In this paper we investigate the source of this bias, seeking to understand its root cause(s) so that it can be effectively mitigated. We perform a series of classification and segmentation experiments on short-axis cine CMR images acquired from Black and White subjects from the UK Biobank and apply AI interpretability methods to understand the results. In the classification experiments, we found that race can be predicted with high accuracy from the images alone, but less accurately from ground truth segmentations, suggesting that the distributional shift between races, which is often the cause of AI bias, is mostly image-based rather than segmentation-based. The interpretability methods showed that most attention in the classification models was focused on non-heart regions, such as subcutaneous fat. Cropping the images tightly around the heart reduced classification accuracy to around chance level. Similarly, race can be predicted from the latent representations of a biased segmentation model, suggesting that race information is encoded in the model. Cropping images tightly around the heart reduced but did not eliminate segmentation bias. We also investigate the influence of possible confounders on the bias observed.
Uncertainty Aware Training to Improve Deep Learning Model Calibration for Classification of Cardiac MR Images
Aug 29, 2023Abstract:Quantifying uncertainty of predictions has been identified as one way to develop more trustworthy artificial intelligence (AI) models beyond conventional reporting of performance metrics. When considering their role in a clinical decision support setting, AI classification models should ideally avoid confident wrong predictions and maximise the confidence of correct predictions. Models that do this are said to be well-calibrated with regard to confidence. However, relatively little attention has been paid to how to improve calibration when training these models, i.e., to make the training strategy uncertainty-aware. In this work we evaluate three novel uncertainty-aware training strategies comparing against two state-of-the-art approaches. We analyse performance on two different clinical applications: cardiac resynchronisation therapy (CRT) response prediction and coronary artery disease (CAD) diagnosis from cardiac magnetic resonance (CMR) images. The best-performing model in terms of both classification accuracy and the most common calibration measure, expected calibration error (ECE) was the Confidence Weight method, a novel approach that weights the loss of samples to explicitly penalise confident incorrect predictions. The method reduced the ECE by 17% for CRT response prediction and by 22% for CAD diagnosis when compared to a baseline classifier in which no uncertainty-aware strategy was included. In both applications, as well as reducing the ECE there was a slight increase in accuracy from 69% to 70% and 70% to 72% for CRT response prediction and CAD diagnosis respectively. However, our analysis showed a lack of consistency in terms of optimal models when using different calibration measures. This indicates the need for careful consideration of performance metrics when training and selecting models for complex high-risk applications in healthcare.
Deep Learning Framework for Spleen Volume Estimation from 2D Cross-sectional Views
Aug 17, 2023



Abstract:Abnormal spleen enlargement (splenomegaly) is regarded as a clinical indicator for a range of conditions, including liver disease, cancer and blood diseases. While spleen length measured from ultrasound images is a commonly used surrogate for spleen size, spleen volume remains the gold standard metric for assessing splenomegaly and the severity of related clinical conditions. Computed tomography is the main imaging modality for measuring spleen volume, but it is less accessible in areas where there is a high prevalence of splenomegaly (e.g., the Global South). Our objective was to enable automated spleen volume measurement from 2D cross-sectional segmentations, which can be obtained from ultrasound imaging. In this study, we describe a variational autoencoder-based framework to measure spleen volume from single- or dual-view 2D spleen segmentations. We propose and evaluate three volume estimation methods within this framework. We also demonstrate how 95% confidence intervals of volume estimates can be produced to make our method more clinically useful. Our best model achieved mean relative volume accuracies of 86.62% and 92.58% for single- and dual-view segmentations, respectively, surpassing the performance of the clinical standard approach of linear regression using manual measurements and a comparative deep learning-based 2D-3D reconstruction-based approach. The proposed spleen volume estimation framework can be integrated into standard clinical workflows which currently use 2D ultrasound images to measure spleen length. To the best of our knowledge, this is the first work to achieve direct 3D spleen volume estimation from 2D spleen segmentations.
Addressing Deep Learning Model Calibration Using Evidential Neural Networks and Uncertainty-Aware Training
Jan 30, 2023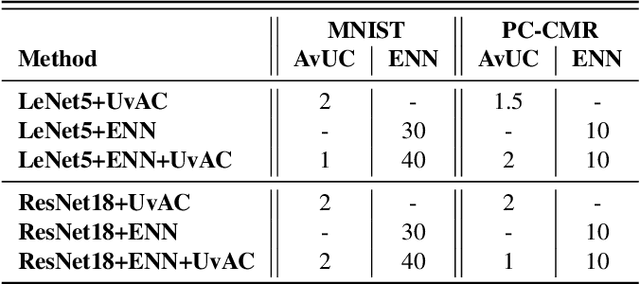

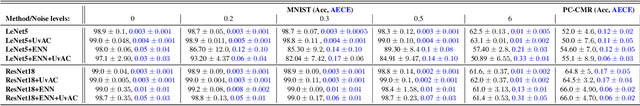
Abstract:In terms of accuracy, deep learning (DL) models have had considerable success in classification problems for medical imaging applications. However, it is well-known that the outputs of such models, which typically utilise the SoftMax function in the final classification layer can be over-confident, i.e. they are poorly calibrated. Two competing solutions to this problem have been proposed: uncertainty-aware training and evidential neural networks (ENNs). In this paper, we perform an investigation into the improvements to model calibration that can be achieved by each of these approaches individually, and their combination. We perform experiments on two classification tasks: a simpler MNIST digit classification task and a more complex and realistic medical imaging artefact detection task using Phase Contrast Cardiac Magnetic Resonance images. The experimental results demonstrate that model calibration can suffer when the task becomes challenging enough to require a higher-capacity model. However, in our complex artefact detection task, we saw an improvement in calibration for both a low and higher-capacity model when implementing both the ENN and uncertainty-aware training together, indicating that this approach can offer a promising way to improve calibration in such settings. The findings highlight the potential use of these approaches to improve model calibration in a complex application, which would in turn improve clinician trust in DL models.
A systematic study of race and sex bias in CNN-based cardiac MR segmentation
Sep 04, 2022
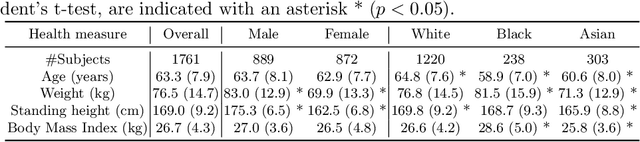


Abstract:In computer vision there has been significant research interest in assessing potential demographic bias in deep learning models. One of the main causes of such bias is imbalance in the training data. In medical imaging, where the potential impact of bias is arguably much greater, there has been less interest. In medical imaging pipelines, segmentation of structures of interest plays an important role in estimating clinical biomarkers that are subsequently used to inform patient management. Convolutional neural networks (CNNs) are starting to be used to automate this process. We present the first systematic study of the impact of training set imbalance on race and sex bias in CNN-based segmentation. We focus on segmentation of the structures of the heart from short axis cine cardiac magnetic resonance images, and train multiple CNN segmentation models with different levels of race/sex imbalance. We find no significant bias in the sex experiment but significant bias in two separate race experiments, highlighting the need to consider adequate representation of different demographic groups in health datasets.
Deep Learning-based Segmentation of Pleural Effusion From Ultrasound Using Coordinate Convolutions
Aug 05, 2022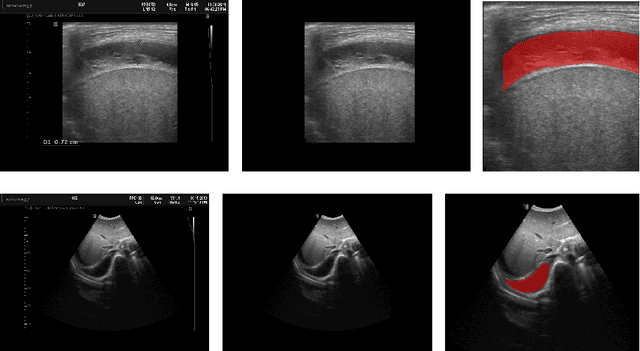

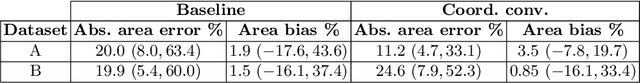
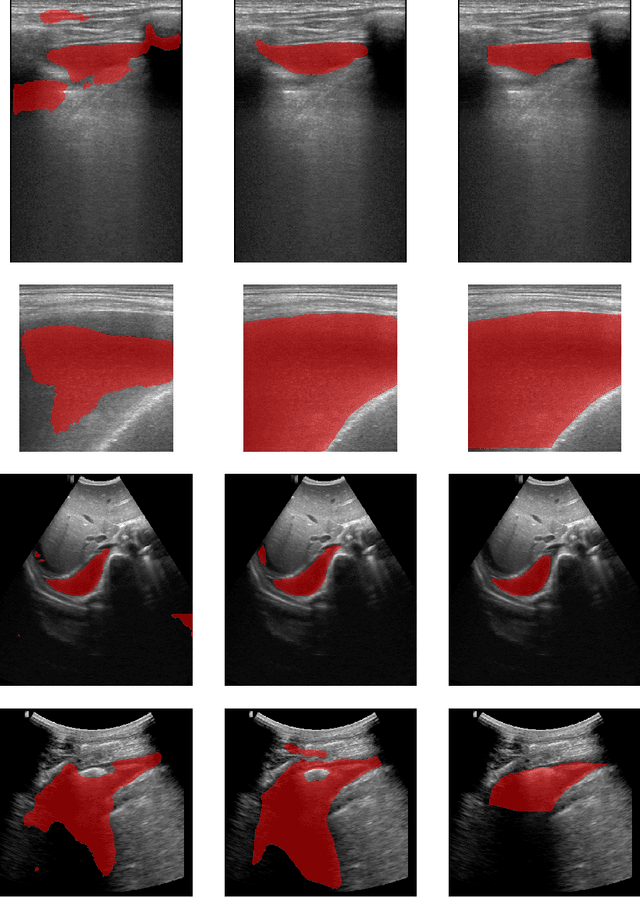
Abstract:In many low-to-middle income (LMIC) countries, ultrasound is used for assessment of pleural effusion. Typically, the extent of the effusion is manually measured by a sonographer, leading to significant intra-/inter-observer variability. In this work, we investigate the use of deep learning (DL) to automate the process of pleural effusion segmentation from ultrasound images. On two datasets acquired in a LMIC setting, we achieve median Dice Similarity Coefficients (DSCs) of 0.82 and 0.74 respectively using the nnU-net DL model. We also investigate the use of coordinate convolutions in the DL model and find that this results in a statistically significant improvement in the median DSC on the first dataset to 0.85, with no significant change on the second dataset. This work showcases, for the first time, the potential of DL in automating the process of effusion assessment from ultrasound in LMIC settings where there is often a lack of experienced radiologists to perform such tasks.
The Impact of Domain Shift on Left and Right Ventricle Segmentation in Short Axis Cardiac MR Images
Sep 22, 2021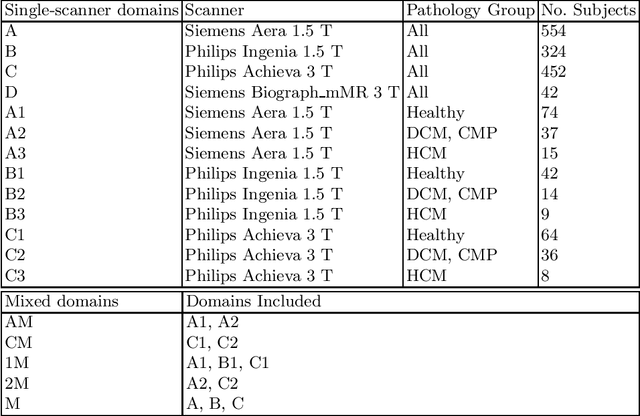
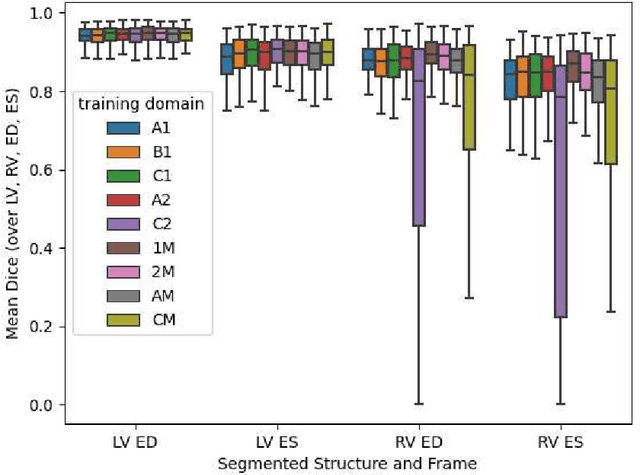
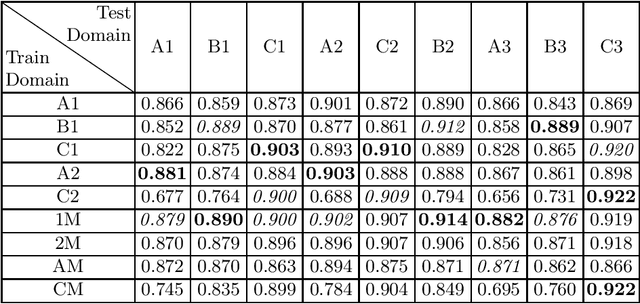

Abstract:Domain shift refers to the difference in the data distribution of two datasets, normally between the training set and the test set for machine learning algorithms. Domain shift is a serious problem for generalization of machine learning models and it is well-established that a domain shift between the training and test sets may cause a drastic drop in the model's performance. In medical imaging, there can be many sources of domain shift such as different scanners or scan protocols, different pathologies in the patient population, anatomical differences in the patient population (e.g. men vs women) etc. Therefore, in order to train models that have good generalization performance, it is important to be aware of the domain shift problem, its potential causes and to devise ways to address it. In this paper, we study the effect of domain shift on left and right ventricle blood pool segmentation in short axis cardiac MR images. Our dataset contains short axis images from 4 different MR scanners and 3 different pathology groups. The training is performed with nnUNet. The results show that scanner differences cause a greater drop in performance compared to changing the pathology group, and that the impact of domain shift is greater on right ventricle segmentation compared to left ventricle segmentation. Increasing the number of training subjects increased cross-scanner performance more than in-scanner performance at small training set sizes, but this difference in improvement decreased with larger training set sizes. Training models using data from multiple scanners improved cross-domain performance.
Quality-aware Cine Cardiac MRI Reconstruction and Analysis from Undersampled k-space Data
Sep 16, 2021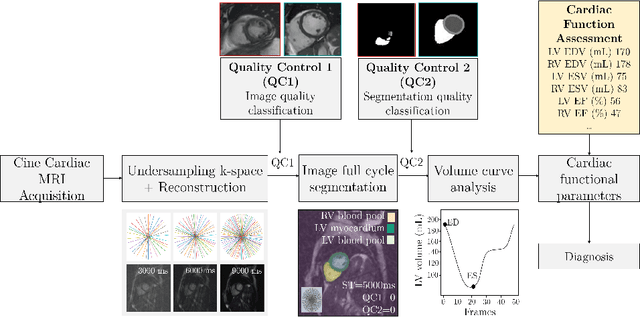
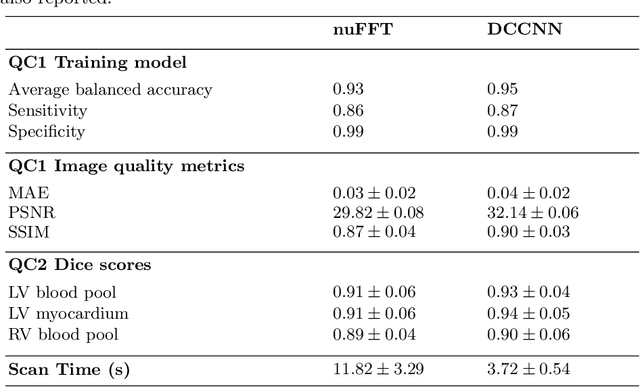
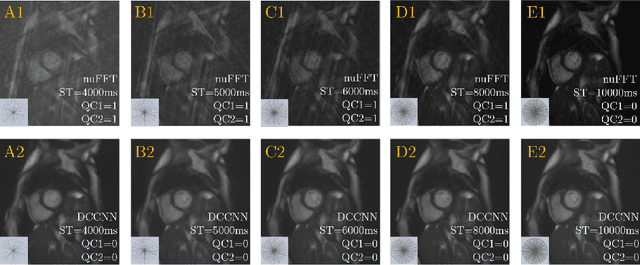
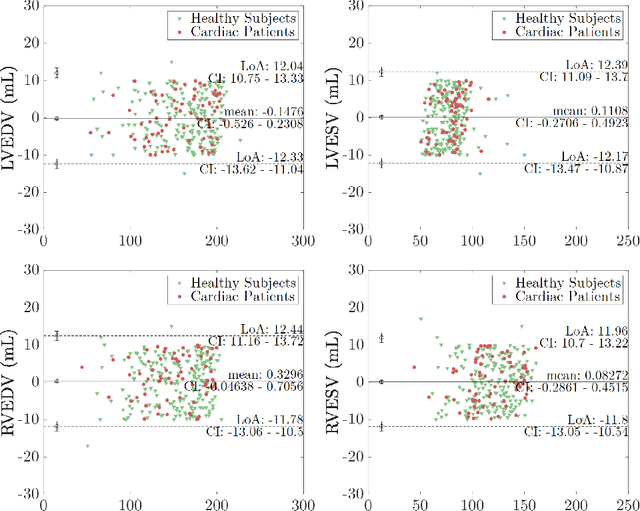
Abstract:Cine cardiac MRI is routinely acquired for the assessment of cardiac health, but the imaging process is slow and typically requires several breath-holds to acquire sufficient k-space profiles to ensure good image quality. Several undersampling-based reconstruction techniques have been proposed during the last decades to speed up cine cardiac MRI acquisition. However, the undersampling factor is commonly fixed to conservative values before acquisition to ensure diagnostic image quality, potentially leading to unnecessarily long scan times. In this paper, we propose an end-to-end quality-aware cine short-axis cardiac MRI framework that combines image acquisition and reconstruction with downstream tasks such as segmentation, volume curve analysis and estimation of cardiac functional parameters. The goal is to reduce scan time by acquiring only a fraction of k-space data to enable the reconstruction of images that can pass quality control checks and produce reliable estimates of cardiac functional parameters. The framework consists of a deep learning model for the reconstruction of 2D+t cardiac cine MRI images from undersampled data, an image quality-control step to detect good quality reconstructions, followed by a deep learning model for bi-ventricular segmentation, a quality-control step to detect good quality segmentations and automated calculation of cardiac functional parameters. To demonstrate the feasibility of the proposed approach, we perform simulations using a cohort of selected participants from the UK Biobank (n=270), 200 healthy subjects and 70 patients with cardiomyopathies. Our results show that we can produce quality-controlled images in a scan time reduced from 12 to 4 seconds per slice, enabling reliable estimates of cardiac functional parameters such as ejection fraction within 5% mean absolute error.
Fairness in Cardiac MR Image Analysis: An Investigation of Bias Due to Data Imbalance in Deep Learning Based Segmentation
Jul 01, 2021
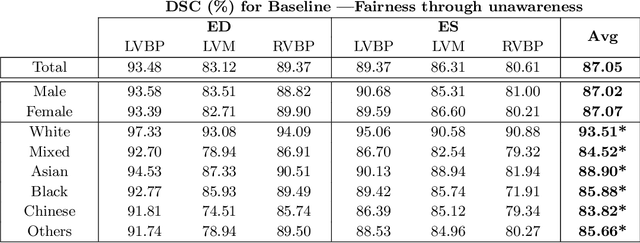

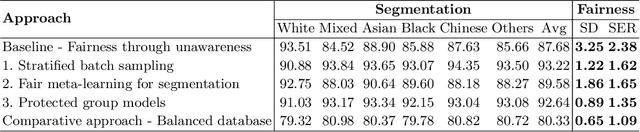
Abstract:The subject of "fairness" in artificial intelligence (AI) refers to assessing AI algorithms for potential bias based on demographic characteristics such as race and gender, and the development of algorithms to address this bias. Most applications to date have been in computer vision, although some work in healthcare has started to emerge. The use of deep learning (DL) in cardiac MR segmentation has led to impressive results in recent years, and such techniques are starting to be translated into clinical practice. However, no work has yet investigated the fairness of such models. In this work, we perform such an analysis for racial/gender groups, focusing on the problem of training data imbalance, using a nnU-Net model trained and evaluated on cine short axis cardiac MR data from the UK Biobank dataset, consisting of 5,903 subjects from 6 different racial groups. We find statistically significant differences in Dice performance between different racial groups. To reduce the racial bias, we investigated three strategies: (1) stratified batch sampling, in which batch sampling is stratified to ensure balance between racial groups; (2) fair meta-learning for segmentation, in which a DL classifier is trained to classify race and jointly optimized with the segmentation model; and (3) protected group models, in which a different segmentation model is trained for each racial group. We also compared the results to the scenario where we have a perfectly balanced database. To assess fairness we used the standard deviation (SD) and skewed error ratio (SER) of the average Dice values. Our results demonstrate that the racial bias results from the use of imbalanced training data, and that all proposed bias mitigation strategies improved fairness, with the best SD and SER resulting from the use of protected group models.
Deep Learning for Automatic Spleen Length Measurement in Sickle Cell Disease Patients
Sep 06, 2020
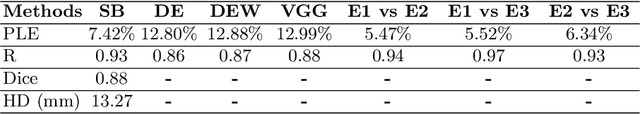
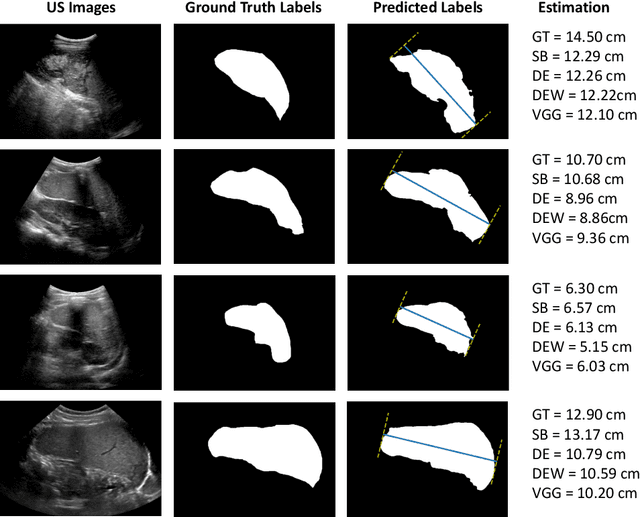
Abstract:Sickle Cell Disease (SCD) is one of the most common genetic diseases in the world. Splenomegaly (abnormal enlargement of the spleen) is frequent among children with SCD. If left untreated, splenomegaly can be life-threatening. The current workflow to measure spleen size includes palpation, possibly followed by manual length measurement in 2D ultrasound imaging. However, this manual measurement is dependent on operator expertise and is subject to intra- and inter-observer variability. We investigate the use of deep learning to perform automatic estimation of spleen length from ultrasound images. We investigate two types of approach, one segmentation-based and one based on direct length estimation, and compare the results against measurements made by human experts. Our best model (segmentation-based) achieved a percentage length error of 7.42%, which is approaching the level of inter-observer variability (5.47%-6.34%). To the best of our knowledge, this is the first attempt to measure spleen size in a fully automated way from ultrasound images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge