Bradley Porter
Uncertainty Aware Training to Improve Deep Learning Model Calibration for Classification of Cardiac MR Images
Aug 29, 2023Abstract:Quantifying uncertainty of predictions has been identified as one way to develop more trustworthy artificial intelligence (AI) models beyond conventional reporting of performance metrics. When considering their role in a clinical decision support setting, AI classification models should ideally avoid confident wrong predictions and maximise the confidence of correct predictions. Models that do this are said to be well-calibrated with regard to confidence. However, relatively little attention has been paid to how to improve calibration when training these models, i.e., to make the training strategy uncertainty-aware. In this work we evaluate three novel uncertainty-aware training strategies comparing against two state-of-the-art approaches. We analyse performance on two different clinical applications: cardiac resynchronisation therapy (CRT) response prediction and coronary artery disease (CAD) diagnosis from cardiac magnetic resonance (CMR) images. The best-performing model in terms of both classification accuracy and the most common calibration measure, expected calibration error (ECE) was the Confidence Weight method, a novel approach that weights the loss of samples to explicitly penalise confident incorrect predictions. The method reduced the ECE by 17% for CRT response prediction and by 22% for CAD diagnosis when compared to a baseline classifier in which no uncertainty-aware strategy was included. In both applications, as well as reducing the ECE there was a slight increase in accuracy from 69% to 70% and 70% to 72% for CRT response prediction and CAD diagnosis respectively. However, our analysis showed a lack of consistency in terms of optimal models when using different calibration measures. This indicates the need for careful consideration of performance metrics when training and selecting models for complex high-risk applications in healthcare.
AI-enabled Assessment of Cardiac Systolic and Diastolic Function from Echocardiography
Mar 21, 2022
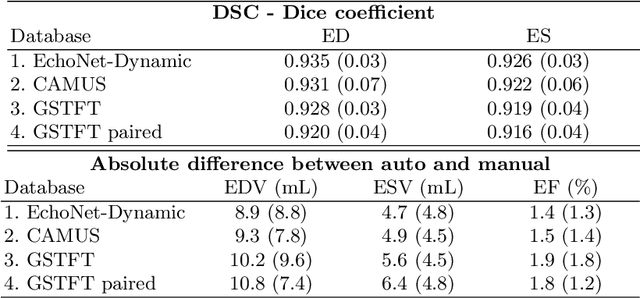
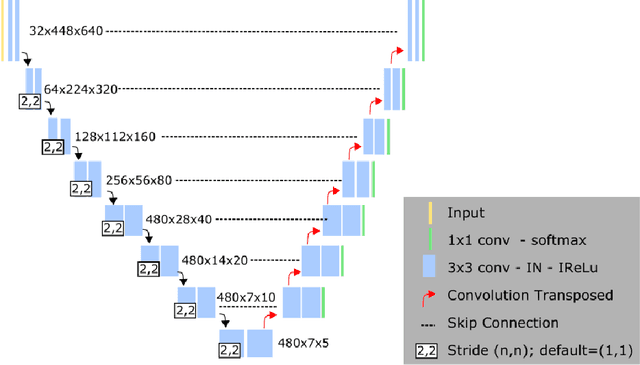
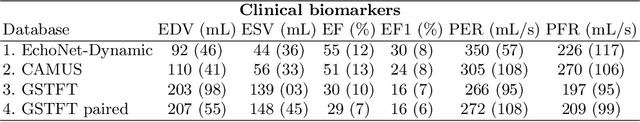
Abstract:Left ventricular (LV) function is an important factor in terms of patient management, outcome, and long-term survival of patients with heart disease. The most recently published clinical guidelines for heart failure recognise that over reliance on only one measure of cardiac function (LV ejection fraction) as a diagnostic and treatment stratification biomarker is suboptimal. Recent advances in AI-based echocardiography analysis have shown excellent results on automated estimation of LV volumes and LV ejection fraction. However, from time-varying 2-D echocardiography acquisition, a richer description of cardiac function can be obtained by estimating functional biomarkers from the complete cardiac cycle. In this work we propose for the first time an AI approach for deriving advanced biomarkers of systolic and diastolic LV function from 2-D echocardiography based on segmentations of the full cardiac cycle. These biomarkers will allow clinicians to obtain a much richer picture of the heart in health and disease. The AI model is based on the 'nn-Unet' framework and was trained and tested using four different databases. Results show excellent agreement between manual and automated analysis and showcase the potential of the advanced systolic and diastolic biomarkers for patient stratification. Finally, for a subset of 50 cases, we perform a correlation analysis between clinical biomarkers derived from echocardiography and CMR and we show excellent agreement between the two modalities.
Uncertainty-Aware Training for Cardiac Resynchronisation Therapy Response Prediction
Sep 22, 2021


Abstract:Evaluation of predictive deep learning (DL) models beyond conventional performance metrics has become increasingly important for applications in sensitive environments like healthcare. Such models might have the capability to encode and analyse large sets of data but they often lack comprehensive interpretability methods, preventing clinical trust in predictive outcomes. Quantifying uncertainty of a prediction is one way to provide such interpretability and promote trust. However, relatively little attention has been paid to how to include such requirements into the training of the model. In this paper we: (i) quantify the data (aleatoric) and model (epistemic) uncertainty of a DL model for Cardiac Resynchronisation Therapy response prediction from cardiac magnetic resonance images, and (ii) propose and perform a preliminary investigation of an uncertainty-aware loss function that can be used to retrain an existing DL image-based classification model to encourage confidence in correct predictions and reduce confidence in incorrect predictions. Our initial results are promising, showing a significant increase in the (epistemic) confidence of true positive predictions, with some evidence of a reduction in false negative confidence.
A Multimodal Deep Learning Model for Cardiac Resynchronisation Therapy Response Prediction
Jul 20, 2021



Abstract:We present a novel multimodal deep learning framework for cardiac resynchronisation therapy (CRT) response prediction from 2D echocardiography and cardiac magnetic resonance (CMR) data. The proposed method first uses the `nnU-Net' segmentation model to extract segmentations of the heart over the full cardiac cycle from the two modalities. Next, a multimodal deep learning classifier is used for CRT response prediction, which combines the latent spaces of the segmentation models of the two modalities. At inference time, this framework can be used with 2D echocardiography data only, whilst taking advantage of the implicit relationship between CMR and echocardiography features learnt from the model. We evaluate our pipeline on a cohort of 50 CRT patients for whom paired echocardiography/CMR data were available, and results show that the proposed multimodal classifier results in a statistically significant improvement in accuracy compared to the baseline approach that uses only 2D echocardiography data. The combination of multimodal data enables CRT response to be predicted with 77.38% accuracy (83.33% sensitivity and 71.43% specificity), which is comparable with the current state-of-the-art in machine learning-based CRT response prediction. Our work represents the first multimodal deep learning approach for CRT response prediction.
Interpretable Deep Models for Cardiac Resynchronisation Therapy Response Prediction
Jul 09, 2020



Abstract:Advances in deep learning (DL) have resulted in impressive accuracy in some medical image classification tasks, but often deep models lack interpretability. The ability of these models to explain their decisions is important for fostering clinical trust and facilitating clinical translation. Furthermore, for many problems in medicine there is a wealth of existing clinical knowledge to draw upon, which may be useful in generating explanations, but it is not obvious how this knowledge can be encoded into DL models - most models are learnt either from scratch or using transfer learning from a different domain. In this paper we address both of these issues. We propose a novel DL framework for image-based classification based on a variational autoencoder (VAE). The framework allows prediction of the output of interest from the latent space of the autoencoder, as well as visualisation (in the image domain) of the effects of crossing the decision boundary, thus enhancing the interpretability of the classifier. Our key contribution is that the VAE disentangles the latent space based on `explanations' drawn from existing clinical knowledge. The framework can predict outputs as well as explanations for these outputs, and also raises the possibility of discovering new biomarkers that are separate (or disentangled) from the existing knowledge. We demonstrate our framework on the problem of predicting response of patients with cardiomyopathy to cardiac resynchronization therapy (CRT) from cine cardiac magnetic resonance images. The sensitivity and specificity of the proposed model on the task of CRT response prediction are 88.43% and 84.39% respectively, and we showcase the potential of our model in enhancing understanding of the factors contributing to CRT response.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge