Naomi Kifle
Deep Learning-based Segmentation of Pleural Effusion From Ultrasound Using Coordinate Convolutions
Aug 05, 2022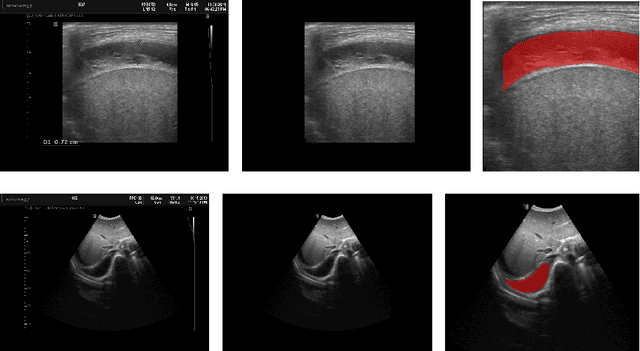

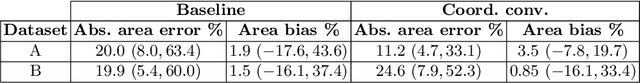
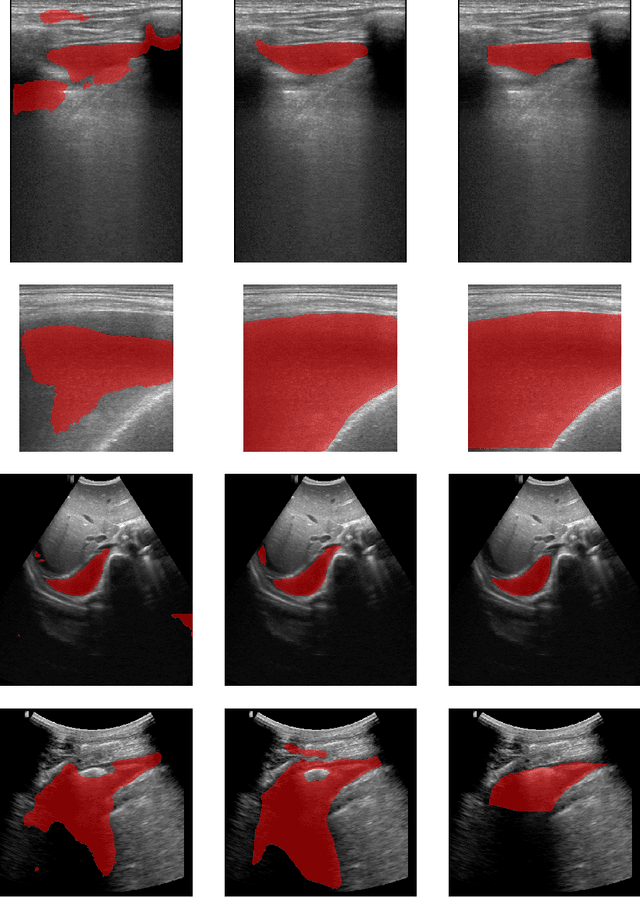
Abstract:In many low-to-middle income (LMIC) countries, ultrasound is used for assessment of pleural effusion. Typically, the extent of the effusion is manually measured by a sonographer, leading to significant intra-/inter-observer variability. In this work, we investigate the use of deep learning (DL) to automate the process of pleural effusion segmentation from ultrasound images. On two datasets acquired in a LMIC setting, we achieve median Dice Similarity Coefficients (DSCs) of 0.82 and 0.74 respectively using the nnU-net DL model. We also investigate the use of coordinate convolutions in the DL model and find that this results in a statistically significant improvement in the median DSC on the first dataset to 0.85, with no significant change on the second dataset. This work showcases, for the first time, the potential of DL in automating the process of effusion assessment from ultrasound in LMIC settings where there is often a lack of experienced radiologists to perform such tasks.
Improved AI-based segmentation of apical and basal slices from clinical cine CMR
Sep 20, 2021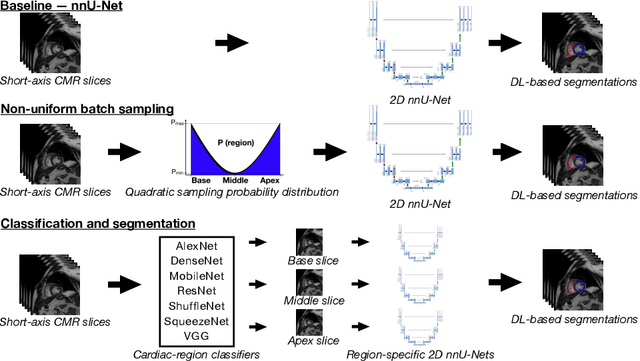



Abstract:Current artificial intelligence (AI) algorithms for short-axis cardiac magnetic resonance (CMR) segmentation achieve human performance for slices situated in the middle of the heart. However, an often-overlooked fact is that segmentation of the basal and apical slices is more difficult. During manual analysis, differences in the basal segmentations have been reported as one of the major sources of disagreement in human interobserver variability. In this work, we aim to investigate the performance of AI algorithms in segmenting basal and apical slices and design strategies to improve their segmentation. We trained all our models on a large dataset of clinical CMR studies obtained from two NHS hospitals (n=4,228) and evaluated them against two external datasets: ACDC (n=100) and M&Ms (n=321). Using manual segmentations as a reference, CMR slices were assigned to one of four regions: non-cardiac, base, middle, and apex. Using the nnU-Net framework as a baseline, we investigated two different approaches to reduce the segmentation performance gap between cardiac regions: (1) non-uniform batch sampling, which allows us to choose how often images from different regions are seen during training; and (2) a cardiac-region classification model followed by three (i.e. base, middle, and apex) region-specific segmentation models. We show that the classification and segmentation approach was best at reducing the performance gap across all datasets. We also show that improvements in the classification performance can subsequently lead to a significantly better performance in the segmentation task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge