Reza Razavi
School of Biomedical Engineering and Imaging Sciences, King's College London, London, UK, Department of Adult and Paediatric Cardiology, Guy's and St Thomas' NHS Foundation Trust, London, UK
Cardiac Digital Twins at Scale from MRI: Open Tools and Representative Models from ~55000 UK Biobank Participants
May 27, 2025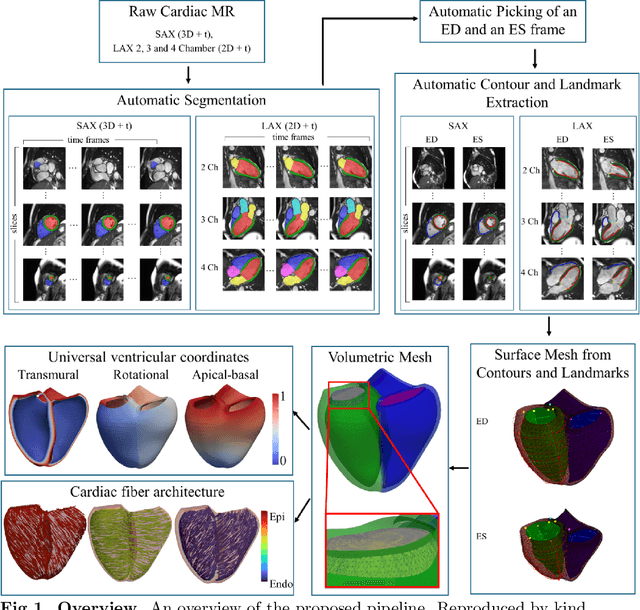
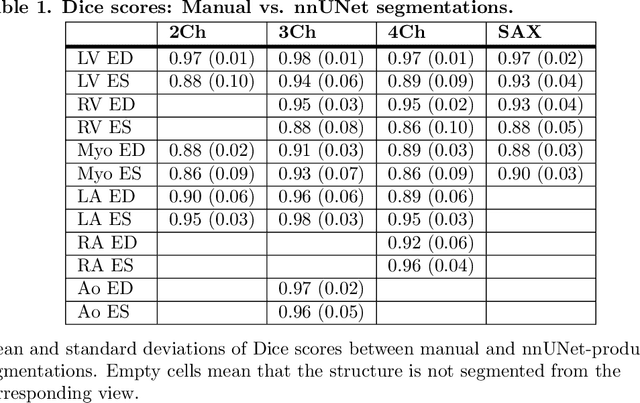
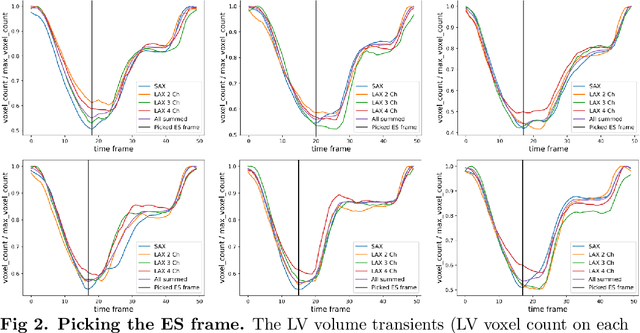
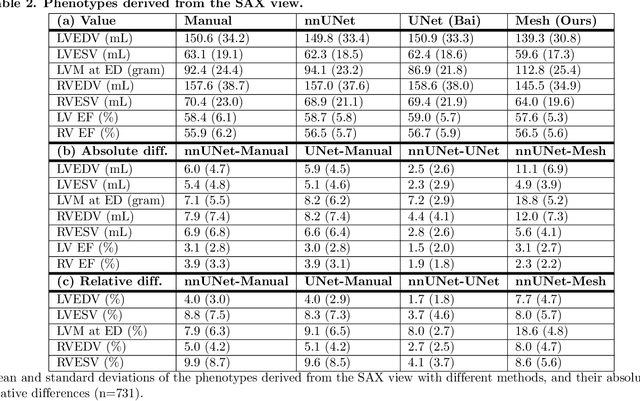
Abstract:A cardiac digital twin is a virtual replica of a patient's heart for screening, diagnosis, prognosis, risk assessment, and treatment planning of cardiovascular diseases. This requires an anatomically accurate patient-specific 3D structural representation of the heart, suitable for electro-mechanical simulations or study of disease mechanisms. However, generation of cardiac digital twins at scale is demanding and there are no public repositories of models across demographic groups. We describe an automatic open-source pipeline for creating patient-specific left and right ventricular meshes from cardiovascular magnetic resonance images, its application to a large cohort of ~55000 participants from UK Biobank, and the construction of the most comprehensive cohort of adult heart models to date, comprising 1423 representative meshes across sex (male, female), body mass index (range: 16 - 42 kg/m$^2$) and age (range: 49 - 80 years). Our code is available at https://github.com/cdttk/biv-volumetric-meshing/tree/plos2025 , and pre-trained networks, representative volumetric meshes with fibers and UVCs will be made available soon.
FedVSR: Towards Model-Agnostic Federated Learning in Video Super-Resolution
Mar 17, 2025Abstract:Video Super-Resolution (VSR) reconstructs high-resolution videos from low-resolution inputs to restore fine details and improve visual clarity. While deep learning-based VSR methods achieve impressive results, their centralized nature raises serious privacy concerns, particularly in applications with strict privacy requirements. Federated Learning (FL) offers an alternative approach, but existing FL methods struggle with low-level vision tasks, leading to suboptimal reconstructions. To address this, we propose FedVSR1, a novel, architecture-independent, and stateless FL framework for VSR. Our approach introduces a lightweight loss term that improves local optimization and guides global aggregation with minimal computational overhead. To the best of our knowledge, this is the first attempt at federated VSR. Extensive experiments show that FedVSR outperforms general FL methods by an average of 0.85 dB in PSNR, highlighting its effectiveness. The code is available at: https://github.com/alimd94/FedVSR
L-FUSION: Laplacian Fetal Ultrasound Segmentation & Uncertainty Estimation
Mar 07, 2025

Abstract:Accurate analysis of prenatal ultrasound (US) is essential for early detection of developmental anomalies. However, operator dependency and technical limitations (e.g. intrinsic artefacts and effects, setting errors) can complicate image interpretation and the assessment of diagnostic uncertainty. We present L-FUSION (Laplacian Fetal US Segmentation with Integrated FoundatiON models), a framework that integrates uncertainty quantification through unsupervised, normative learning and large-scale foundation models for robust segmentation of fetal structures in normal and pathological scans. We propose to utilise the aleatoric logit distributions of Stochastic Segmentation Networks and Laplace approximations with fast Hessian estimations to estimate epistemic uncertainty only from the segmentation head. This enables us to achieve reliable abnormality quantification for instant diagnostic feedback. Combined with an integrated Dropout component, L-FUSION enables reliable differentiation of lesions from normal fetal anatomy with enhanced uncertainty maps and segmentation counterfactuals in US imaging. It improves epistemic and aleatoric uncertainty interpretation and removes the need for manual disease-labelling. Evaluations across multiple datasets show that L-FUSION achieves superior segmentation accuracy and consistent uncertainty quantification, supporting on-site decision-making and offering a scalable solution for advancing fetal ultrasound analysis in clinical settings.
Reversing the Damage: A QP-Aware Transformer-Diffusion Approach for 8K Video Restoration under Codec Compression
Dec 12, 2024Abstract:In this paper, we introduce DiQP; a novel Transformer-Diffusion model for restoring 8K video quality degraded by codec compression. To the best of our knowledge, our model is the first to consider restoring the artifacts introduced by various codecs (AV1, HEVC) by Denoising Diffusion without considering additional noise. This approach allows us to model the complex, non-Gaussian nature of compression artifacts, effectively learning to reverse the degradation. Our architecture combines the power of Transformers to capture long-range dependencies with an enhanced windowed mechanism that preserves spatiotemporal context within groups of pixels across frames. To further enhance restoration, the model incorporates auxiliary "Look Ahead" and "Look Around" modules, providing both future and surrounding frame information to aid in reconstructing fine details and enhancing overall visual quality. Extensive experiments on different datasets demonstrate that our model outperforms state-of-the-art methods, particularly for high-resolution videos such as 4K and 8K, showcasing its effectiveness in restoring perceptually pleasing videos from highly compressed sources.
Improving the Scan-rescan Precision of AI-based CMR Biomarker Estimation
Aug 21, 2024Abstract:Quantification of cardiac biomarkers from cine cardiovascular magnetic resonance (CMR) data using deep learning (DL) methods offers many advantages, such as increased accuracy and faster analysis. However, only a few studies have focused on the scan-rescan precision of the biomarker estimates, which is important for reproducibility and longitudinal analysis. Here, we propose a cardiac biomarker estimation pipeline that not only focuses on achieving high segmentation accuracy but also on improving the scan-rescan precision of the computed biomarkers, namely left and right ventricular ejection fraction, and left ventricular myocardial mass. We evaluate two approaches to improve the apical-basal resolution of the segmentations used for estimating the biomarkers: one based on image interpolation and one based on segmentation interpolation. Using a database comprising scan-rescan cine CMR data acquired from 92 subjects, we compare the performance of these two methods against ground truth (GT) segmentations and DL segmentations obtained before interpolation (baseline). The results demonstrate that both the image-based and segmentation-based interpolation methods were able to narrow Bland-Altman scan-rescan confidence intervals for all biomarkers compared to the GT and baseline performances. Our findings highlight the importance of focusing not only on segmentation accuracy but also on the consistency of biomarkers across repeated scans, which is crucial for longitudinal analysis of cardiac function.
Improving Deep Learning Model Calibration for Cardiac Applications using Deterministic Uncertainty Networks and Uncertainty-aware Training
May 10, 2024



Abstract:Improving calibration performance in deep learning (DL) classification models is important when planning the use of DL in a decision-support setting. In such a scenario, a confident wrong prediction could lead to a lack of trust and/or harm in a high-risk application. We evaluate the impact on accuracy and calibration of two types of approach that aim to improve DL classification model calibration: deterministic uncertainty methods (DUM) and uncertainty-aware training. Specifically, we test the performance of three DUMs and two uncertainty-aware training approaches as well as their combinations. To evaluate their utility, we use two realistic clinical applications from the field of cardiac imaging: artefact detection from phase contrast cardiac magnetic resonance (CMR) and disease diagnosis from the public ACDC CMR dataset. Our results indicate that both DUMs and uncertainty-aware training can improve both accuracy and calibration in both of our applications, with DUMs generally offering the best improvements. We also investigate the combination of the two approaches, resulting in a novel deterministic uncertainty-aware training approach. This provides further improvements for some combinations of DUMs and uncertainty-aware training approaches.
Whole-examination AI estimation of fetal biometrics from 20-week ultrasound scans
Jan 02, 2024Abstract:The current approach to fetal anomaly screening is based on biometric measurements derived from individually selected ultrasound images. In this paper, we introduce a paradigm shift that attains human-level performance in biometric measurement by aggregating automatically extracted biometrics from every frame across an entire scan, with no need for operator intervention. We use a convolutional neural network to classify each frame of an ultrasound video recording. We then measure fetal biometrics in every frame where appropriate anatomy is visible. We use a Bayesian method to estimate the true value of each biometric from a large number of measurements and probabilistically reject outliers. We performed a retrospective experiment on 1457 recordings (comprising 48 million frames) of 20-week ultrasound scans, estimated fetal biometrics in those scans and compared our estimates to the measurements sonographers took during the scan. Our method achieves human-level performance in estimating fetal biometrics and estimates well-calibrated credible intervals in which the true biometric value is expected to lie.
Uncertainty Aware Training to Improve Deep Learning Model Calibration for Classification of Cardiac MR Images
Aug 29, 2023Abstract:Quantifying uncertainty of predictions has been identified as one way to develop more trustworthy artificial intelligence (AI) models beyond conventional reporting of performance metrics. When considering their role in a clinical decision support setting, AI classification models should ideally avoid confident wrong predictions and maximise the confidence of correct predictions. Models that do this are said to be well-calibrated with regard to confidence. However, relatively little attention has been paid to how to improve calibration when training these models, i.e., to make the training strategy uncertainty-aware. In this work we evaluate three novel uncertainty-aware training strategies comparing against two state-of-the-art approaches. We analyse performance on two different clinical applications: cardiac resynchronisation therapy (CRT) response prediction and coronary artery disease (CAD) diagnosis from cardiac magnetic resonance (CMR) images. The best-performing model in terms of both classification accuracy and the most common calibration measure, expected calibration error (ECE) was the Confidence Weight method, a novel approach that weights the loss of samples to explicitly penalise confident incorrect predictions. The method reduced the ECE by 17% for CRT response prediction and by 22% for CAD diagnosis when compared to a baseline classifier in which no uncertainty-aware strategy was included. In both applications, as well as reducing the ECE there was a slight increase in accuracy from 69% to 70% and 70% to 72% for CRT response prediction and CAD diagnosis respectively. However, our analysis showed a lack of consistency in terms of optimal models when using different calibration measures. This indicates the need for careful consideration of performance metrics when training and selecting models for complex high-risk applications in healthcare.
Automatic retrieval of corresponding US views in longitudinal examinations
Jun 07, 2023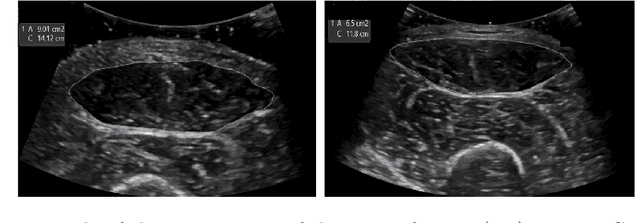
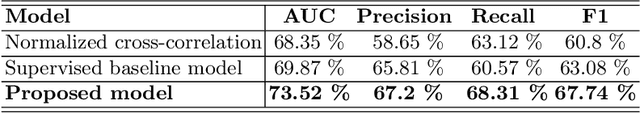
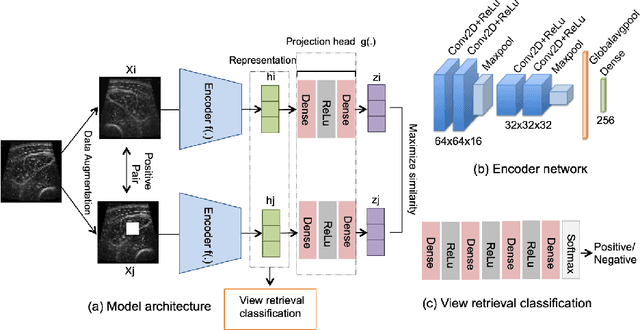
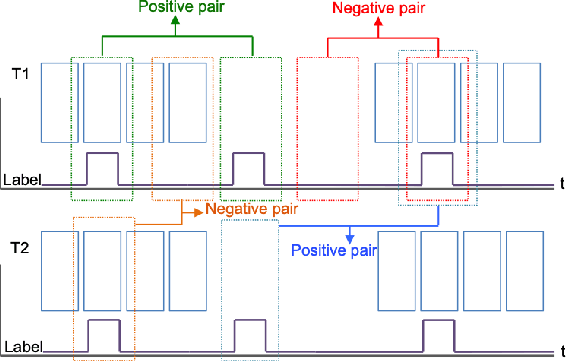
Abstract:Skeletal muscle atrophy is a common occurrence in critically ill patients in the intensive care unit (ICU) who spend long periods in bed. Muscle mass must be recovered through physiotherapy before patient discharge and ultrasound imaging is frequently used to assess the recovery process by measuring the muscle size over time. However, these manual measurements are subject to large variability, particularly since the scans are typically acquired on different days and potentially by different operators. In this paper, we propose a self-supervised contrastive learning approach to automatically retrieve similar ultrasound muscle views at different scan times. Three different models were compared using data from 67 patients acquired in the ICU. Results indicate that our contrastive model outperformed a supervised baseline model in the task of view retrieval with an AUC of 73.52% and when combined with an automatic segmentation model achieved 5.7%+/-0.24% error in cross-sectional area. Furthermore, a user study survey confirmed the efficacy of our model for muscle view retrieval.
Feature-Conditioned Cascaded Video Diffusion Models for Precise Echocardiogram Synthesis
Mar 23, 2023Abstract:Image synthesis is expected to provide value for the translation of machine learning methods into clinical practice. Fundamental problems like model robustness, domain transfer, causal modelling, and operator training become approachable through synthetic data. Especially, heavily operator-dependant modalities like Ultrasound imaging require robust frameworks for image and video generation. So far, video generation has only been possible by providing input data that is as rich as the output data, e.g., image sequence plus conditioning in, video out. However, clinical documentation is usually scarce and only single images are reported and stored, thus retrospective patient-specific analysis or the generation of rich training data becomes impossible with current approaches. In this paper, we extend elucidated diffusion models for video modelling to generate plausible video sequences from single images and arbitrary conditioning with clinical parameters. We explore this idea within the context of echocardiograms by looking into the variation of the Left Ventricle Ejection Fraction, the most essential clinical metric gained from these examinations. We use the publicly available EchoNet-Dynamic dataset for all our experiments. Our image to sequence approach achieves an $R^2$ score of 93%, which is 38 points higher than recently proposed sequence to sequence generation methods. Code and models will be available at: https://github.com/HReynaud/EchoDiffusion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge