Charlene Mauger
Cardiac Digital Twins at Scale from MRI: Open Tools and Representative Models from ~55000 UK Biobank Participants
May 27, 2025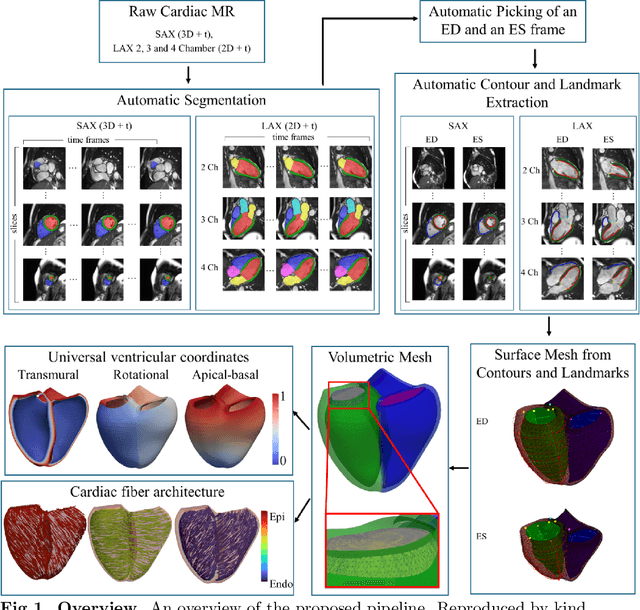
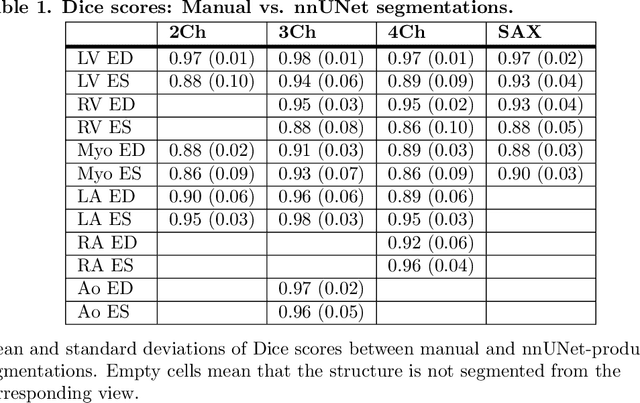
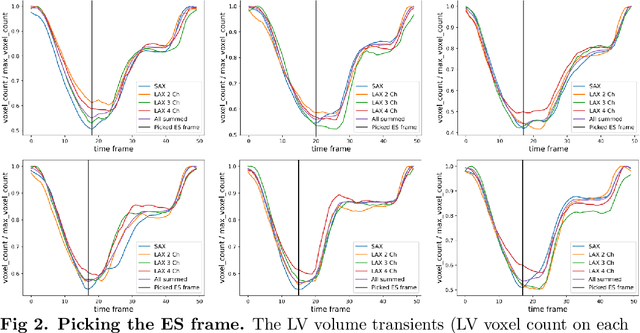
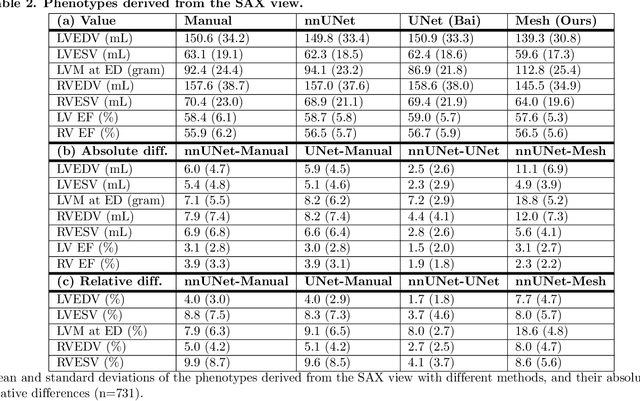
Abstract:A cardiac digital twin is a virtual replica of a patient's heart for screening, diagnosis, prognosis, risk assessment, and treatment planning of cardiovascular diseases. This requires an anatomically accurate patient-specific 3D structural representation of the heart, suitable for electro-mechanical simulations or study of disease mechanisms. However, generation of cardiac digital twins at scale is demanding and there are no public repositories of models across demographic groups. We describe an automatic open-source pipeline for creating patient-specific left and right ventricular meshes from cardiovascular magnetic resonance images, its application to a large cohort of ~55000 participants from UK Biobank, and the construction of the most comprehensive cohort of adult heart models to date, comprising 1423 representative meshes across sex (male, female), body mass index (range: 16 - 42 kg/m$^2$) and age (range: 49 - 80 years). Our code is available at https://github.com/cdttk/biv-volumetric-meshing/tree/plos2025 , and pre-trained networks, representative volumetric meshes with fibers and UVCs will be made available soon.
MorphiNet: A Graph Subdivision Network for Adaptive Bi-ventricle Surface Reconstruction
Dec 14, 2024



Abstract:Cardiac Magnetic Resonance (CMR) imaging is widely used for heart modelling and digital twin computational analysis due to its ability to visualize soft tissues and capture dynamic functions. However, the anisotropic nature of CMR images, characterized by large inter-slice distances and misalignments from cardiac motion, poses significant challenges to accurate model reconstruction. These limitations result in data loss and measurement inaccuracies, hindering the capture of detailed anatomical structures. This study introduces MorphiNet, a novel network that enhances heart model reconstruction by leveraging high-resolution Computer Tomography (CT) images, unpaired with CMR images, to learn heart anatomy. MorphiNet encodes anatomical structures as gradient fields, transforming template meshes into patient-specific geometries. A multi-layer graph subdivision network refines these geometries while maintaining dense point correspondence. The proposed method achieves high anatomy fidelity, demonstrating approximately 40% higher Dice scores, half the Hausdorff distance, and around 3 mm average surface error compared to state-of-the-art methods. MorphiNet delivers superior results with greater inference efficiency. This approach represents a significant advancement in addressing the challenges of CMR-based heart model reconstruction, potentially improving digital twin computational analyses of cardiac structure and functions.
Non-invasive hemodynamic analysis for aortic regurgitation using computational fluid dynamics and deep learning
Nov 23, 2021
Abstract:Changes in cardiovascular hemodynamics are closely related to the development of aortic regurgitation (AR), a type of valvular heart disease. Pressure gradients derived from blood flows are used to indicate AR onset and evaluate its severity. These metrics can be non-invasively obtained using four-dimensional (4D) flow magnetic resonance imaging (MRI), where accuracy is primarily dependent on spatial resolution. However, insufficient resolution often results from limitations in 4D flow MRI and complex AR hemodynamics. To address this, computational fluid dynamics simulations were transformed into synthetic 4D flow MRI data and used to train a variety of neural networks. These networks generated super resolution, full-field phase images with an upsample factor of 4. Results showed decreased velocity error, high structural similarity scores, and improved learning capabilities from previous work. Further validation was performed on two sets of in-vivo 4D flow MRI data and demonstrated success in de-noising flow images. This approach presents an opportunity to comprehensively analyse AR hemodynamics in a non-invasive manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge