David Newby
MorphiNet: A Graph Subdivision Network for Adaptive Bi-ventricle Surface Reconstruction
Dec 14, 2024



Abstract:Cardiac Magnetic Resonance (CMR) imaging is widely used for heart modelling and digital twin computational analysis due to its ability to visualize soft tissues and capture dynamic functions. However, the anisotropic nature of CMR images, characterized by large inter-slice distances and misalignments from cardiac motion, poses significant challenges to accurate model reconstruction. These limitations result in data loss and measurement inaccuracies, hindering the capture of detailed anatomical structures. This study introduces MorphiNet, a novel network that enhances heart model reconstruction by leveraging high-resolution Computer Tomography (CT) images, unpaired with CMR images, to learn heart anatomy. MorphiNet encodes anatomical structures as gradient fields, transforming template meshes into patient-specific geometries. A multi-layer graph subdivision network refines these geometries while maintaining dense point correspondence. The proposed method achieves high anatomy fidelity, demonstrating approximately 40% higher Dice scores, half the Hausdorff distance, and around 3 mm average surface error compared to state-of-the-art methods. MorphiNet delivers superior results with greater inference efficiency. This approach represents a significant advancement in addressing the challenges of CMR-based heart model reconstruction, potentially improving digital twin computational analyses of cardiac structure and functions.
Disentangle, align and fuse for multimodal and zero-shot image segmentation
Nov 12, 2019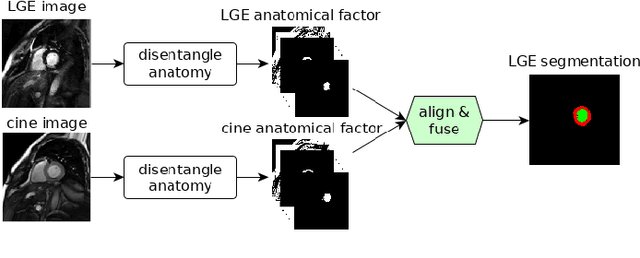
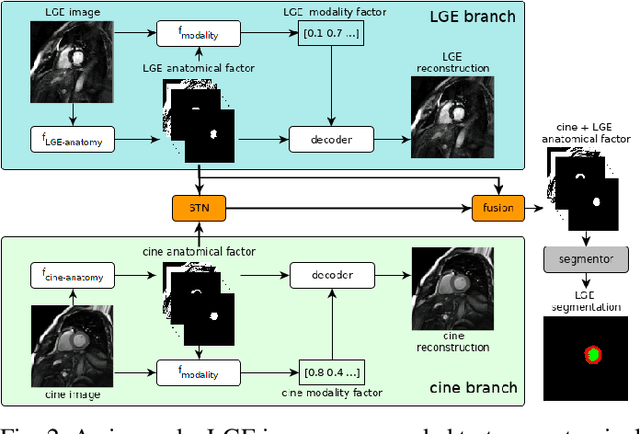
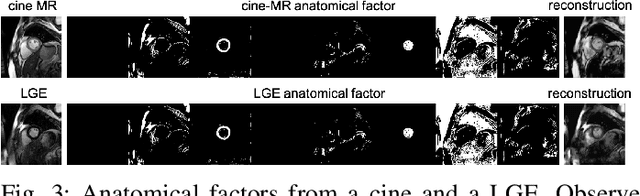
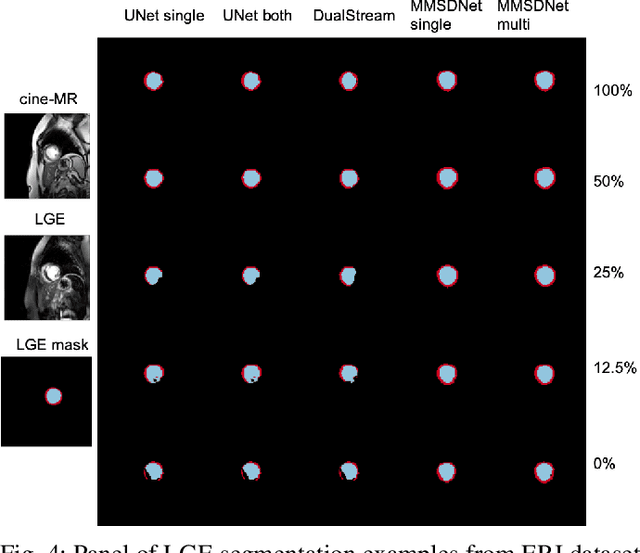
Abstract:Magnetic resonance (MR) protocols rely on several sequences to properly assess pathology and organ status. Yet, despite advances in image analysis we tend to treat each sequence, here termed modality, in isolation. Taking advantage of the information shared between modalities (largely an organ's anatomy) is beneficial for multi-modality multi-input processing and learning. However, we must overcome inherent anatomical misregistrations and disparities in signal intensity across the modalities to claim this benefit. We present a method that offers improved segmentation accuracy of the modality of interest (over a single input model), by learning to leverage information present in other modalities, enabling semi-supervised and zero shot learning. Core to our method is learning a disentangled decomposition into anatomical and imaging factors. Shared anatomical factors from the different inputs are jointly processed and fused to extract more accurate segmentation masks. Image misregistrations are corrected with a Spatial Transformer Network, that non-linearly aligns the anatomical factors. The imaging factor captures signal intensity characteristics across different modality data, and is used for image reconstruction, enabling semi-supervised learning. Temporal and slice pairing between inputs are learned dynamically. We demonstrate applications in Late Gadolinium Enhanced (LGE) and Blood Oxygenation Level Dependent (BOLD) cardiac segmentation, as well as in T2 abdominal segmentation.
Factorised Representation Learning in Cardiac Image Analysis
Mar 22, 2019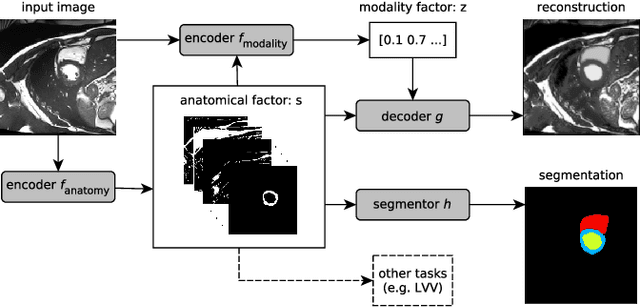
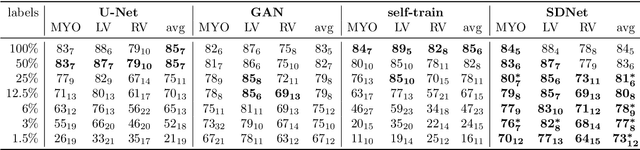
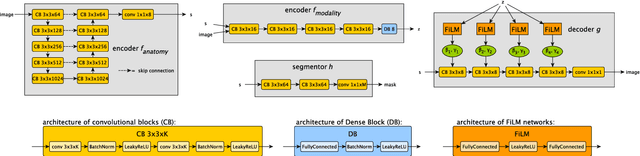
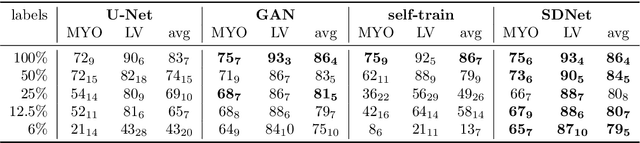
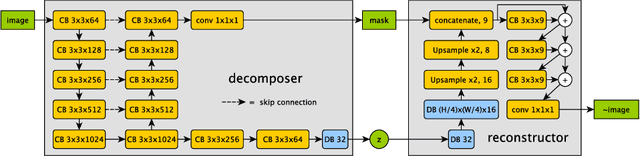
Abstract:Typically, a medical image offers spatial information on the anatomy (and pathology) modulated by imaging specific characteristics. Many imaging modalities including Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) can be interpreted in this way. We can venture further and consider that a medical image naturally factors into some spatial factors depicting anatomy and factors that denote the imaging characteristics. Here, we explicitly learn this decomposed (factorised) representation of imaging data, focusing in particular on cardiac images. We propose Spatial Decomposition Network (SDNet), which factorises 2D medical images into spatial anatomical factors and non-spatial imaging factors. We demonstrate that this high-level representation is ideally suited for several medical image analysis tasks, such as semi-supervised segmentation, multi-task segmentation and regression, and image-to-image synthesis. Specifically, we show that our model can match the performance of fully supervised segmentation models, using only a fraction of the labelled images. Critically, we show that our factorised representation also benefits from supervision obtained either when we use auxiliary tasks to train the model in a multi-task setting (e.g. regressing to known cardiac indices), or when aggregating multimodal data from different sources (e.g. pooling together MRI and CT data). To explore the properties of the learned factorisation, we perform latent-space arithmetic and show that we can synthesise CT from MR and vice versa, by swapping the modality factors. We also demonstrate that the factor holding image specific information can be used to predict the input modality with high accuracy.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019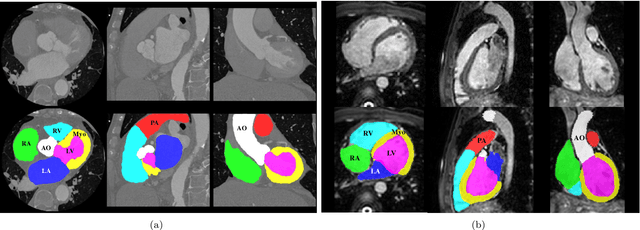
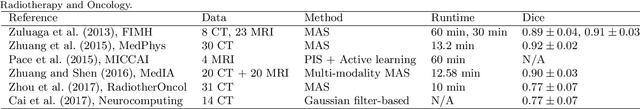
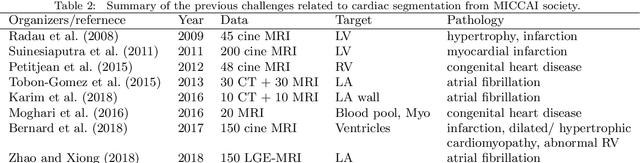
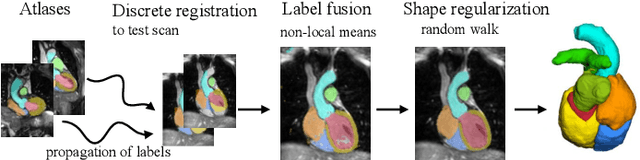
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
Factorised spatial representation learning: application in semi-supervised myocardial segmentation
Nov 02, 2018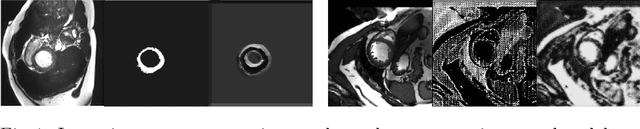
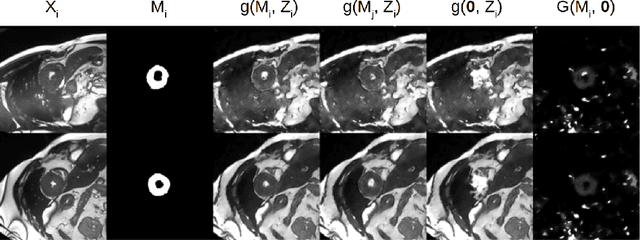
Abstract:The success and generalisation of deep learning algorithms heavily depend on learning good feature representations. In medical imaging this entails representing anatomical information, as well as properties related to the specific imaging setting. Anatomical information is required to perform further analysis, whereas imaging information is key to disentangle scanner variability and potential artefacts. The ability to factorise these would allow for training algorithms only on the relevant information according to the task. To date, such factorisation has not been attempted. In this paper, we propose a methodology of latent space factorisation relying on the cycle-consistency principle. As an example application, we consider cardiac MR segmentation, where we separate information related to the myocardium from other features related to imaging and surrounding substructures. We demonstrate the proposed method's utility in a semi-supervised setting: we use very few labelled images together with many unlabelled images to train a myocardium segmentation neural network. Specifically, we achieve comparable performance to fully supervised networks using a fraction of labelled images in experiments on ACDC and a dataset from Edinburgh Imaging Facility QMRI. Code will be made available at https://github.com/agis85/spatial_factorisation.
Unsupervised learning for cross-domain medical image synthesis using deformation invariant cycle consistency networks
Aug 12, 2018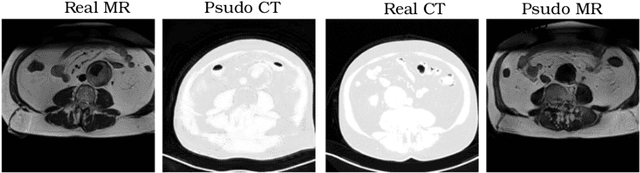
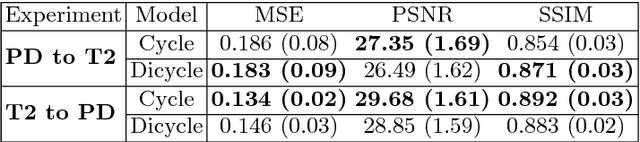
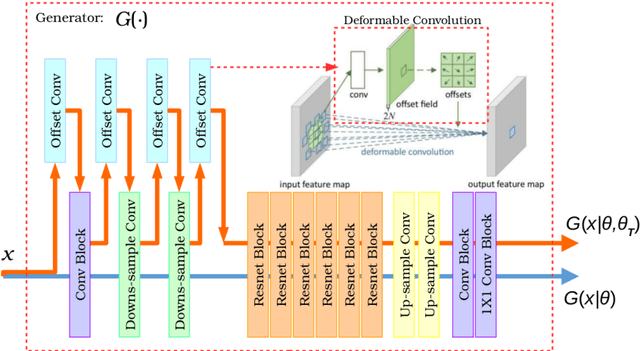
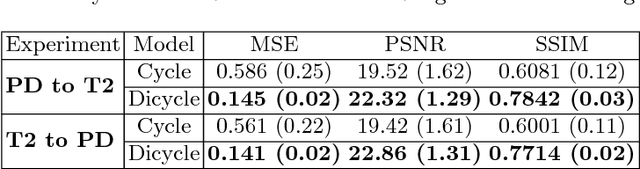
Abstract:Recently, the cycle-consistent generative adversarial networks (CycleGAN) has been widely used for synthesis of multi-domain medical images. The domain-specific nonlinear deformations captured by CycleGAN make the synthesized images difficult to be used for some applications, for example, generating pseudo-CT for PET-MR attenuation correction. This paper presents a deformation-invariant CycleGAN (DicycleGAN) method using deformable convolutional layers and new cycle-consistency losses. Its robustness dealing with data that suffer from domain-specific nonlinear deformations has been evaluated through comparison experiments performed on a multi-sequence brain MR dataset and a multi-modality abdominal dataset. Our method has displayed its ability to generate synthesized data that is aligned with the source while maintaining a proper quality of signal compared to CycleGAN-generated data. The proposed model also obtained comparable performance with CycleGAN when data from the source and target domains are alignable through simple affine transformations.
A two-stage 3D Unet framework for multi-class segmentation on full resolution image
Apr 12, 2018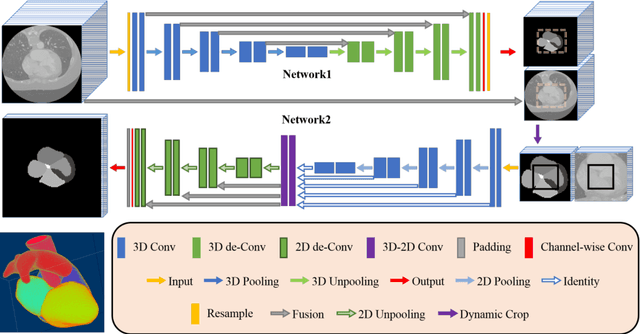
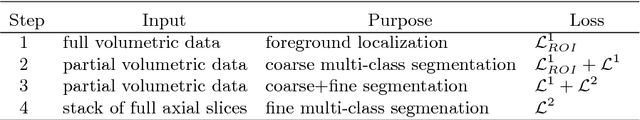
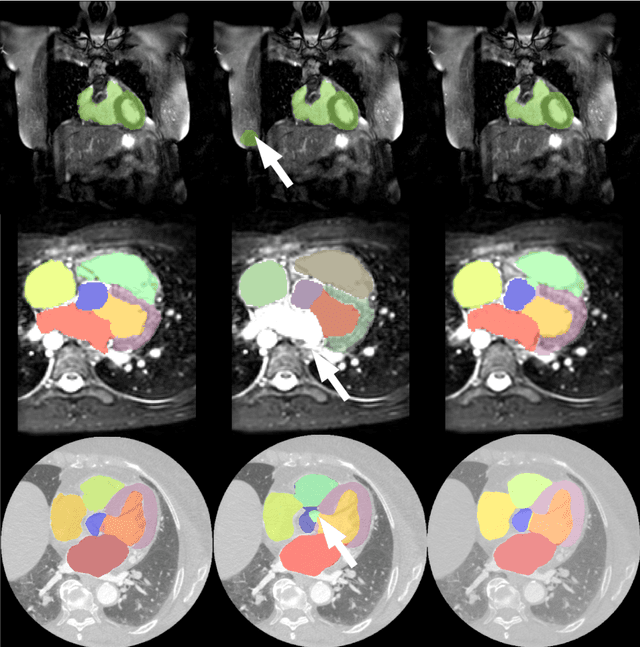
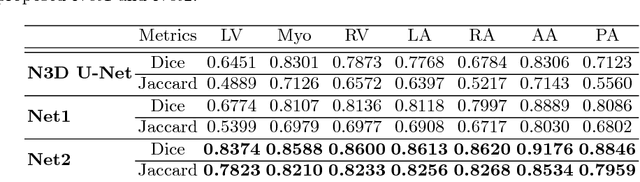
Abstract:Deep convolutional neural networks (CNNs) have been intensively used for multi-class segmentation of data from different modalities and achieved state-of-the-art performances. However, a common problem when dealing with large, high resolution 3D data is that the volumes input into the deep CNNs has to be either cropped or downsampled due to limited memory capacity of computing devices. These operations lead to loss of resolution and increment of class imbalance in the input data batches, which can downgrade the performances of segmentation algorithms. Inspired by the architecture of image super-resolution CNN (SRCNN) and self-normalization network (SNN), we developed a two-stage modified Unet framework that simultaneously learns to detect a ROI within the full volume and to classify voxels without losing the original resolution. Experiments on a variety of multi-modal volumes demonstrated that, when trained with a simply weighted dice coefficients and our customized learning procedure, this framework shows better segmentation performances than state-of-the-art Deep CNNs with advanced similarity metrics.
A Distance Oriented Kalman Filter Particle Swarm Optimizer Applied to Multi-Modality Image Registration
Mar 20, 2018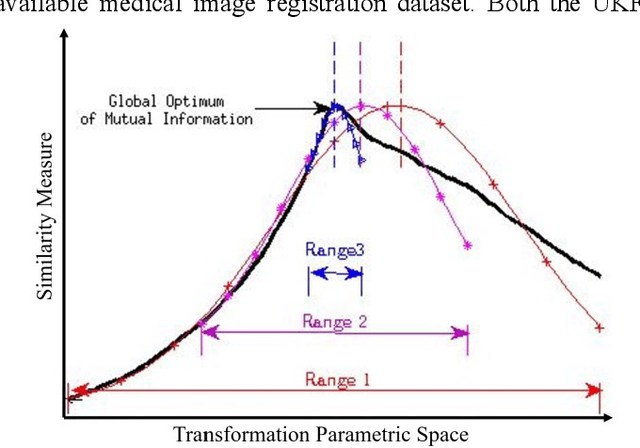
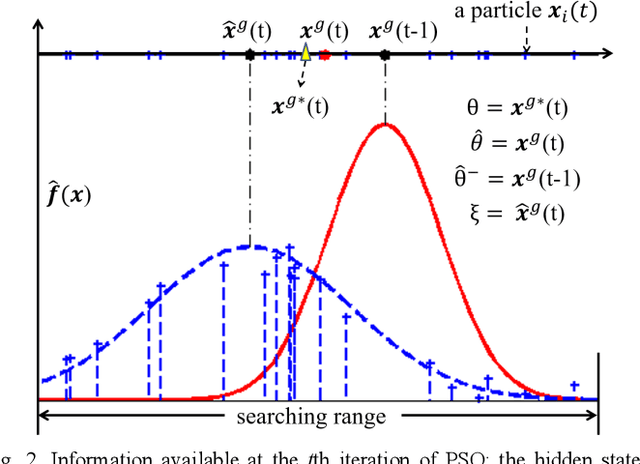
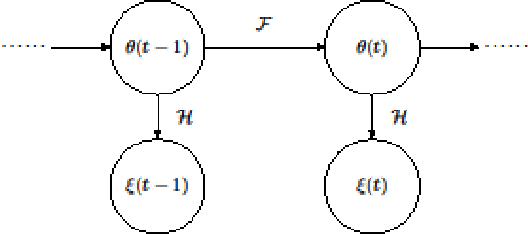
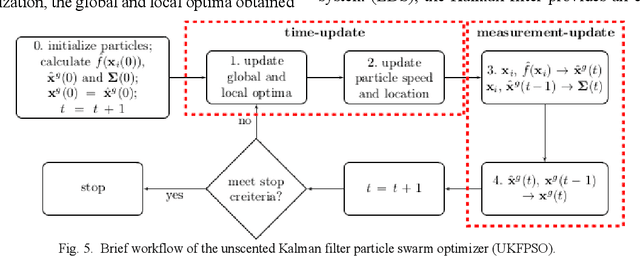
Abstract:In this paper we describe improvements to the particle swarm optimizer (PSO) made by inclusion of an unscented Kalman filter to guide particle motion. We demonstrate the effectiveness of the unscented Kalman filter PSO by comparing it with the original PSO algorithm and its variants designed to improve performance. The PSOs were tested firstly on a number of common synthetic benchmarking functions, and secondly applied to a practical three-dimensional image registration problem. The proposed methods displayed better performances for 4 out of 8 benchmark functions, and reduced the target registration errors by at least 2mm when registering down-sampled benchmark brain images. Our methods also demonstrated an ability to align images featuring motion related artefacts which all other methods failed to register. These new PSO methods provide a novel, efficient mechanism to integrate prior knowledge into each iteration of the optimization process, which can enhance the accuracy and speed of convergence in the application of medical image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge