Agisilaos Chartsias
Uncertainty Propagation for Echocardiography Clinical Metric Estimation via Contour Sampling
Feb 18, 2025Abstract:Echocardiography plays a fundamental role in the extraction of important clinical parameters (e.g. left ventricular volume and ejection fraction) required to determine the presence and severity of heart-related conditions. When deploying automated techniques for computing these parameters, uncertainty estimation is crucial for assessing their utility. Since clinical parameters are usually derived from segmentation maps, there is no clear path for converting pixel-wise uncertainty values into uncertainty estimates in the downstream clinical metric calculation. In this work, we propose a novel uncertainty estimation method based on contouring rather than segmentation. Our method explicitly predicts contour location uncertainty from which contour samples can be drawn. Finally, the sampled contours can be used to propagate uncertainty to clinical metrics. Our proposed method not only provides accurate uncertainty estimations for the task of contouring but also for the downstream clinical metrics on two cardiac ultrasound datasets. Code is available at: https://github.com/ThierryJudge/contouring-uncertainty.
Multi-Site Class-Incremental Learning with Weighted Experts in Echocardiography
Jul 31, 2024Abstract:Building an echocardiography view classifier that maintains performance in real-life cases requires diverse multi-site data, and frequent updates with newly available data to mitigate model drift. Simply fine-tuning on new datasets results in "catastrophic forgetting", and cannot adapt to variations of view labels between sites. Alternatively, collecting all data on a single server and re-training may not be feasible as data sharing agreements may restrict image transfer, or datasets may only become available at different times. Furthermore, time and cost associated with re-training grows with every new dataset. We propose a class-incremental learning method which learns an expert network for each dataset, and combines all expert networks with a score fusion model. The influence of ``unqualified experts'' is minimised by weighting each contribution with a learnt in-distribution score. These weights promote transparency as the contribution of each expert is known during inference. Instead of using the original images, we use learned features from each dataset, which are easier to share and raise fewer licensing and privacy concerns. We validate our work on six datasets from multiple sites, demonstrating significant reductions in training time while improving view classification performance.
BackMix: Mitigating Shortcut Learning in Echocardiography with Minimal Supervision
Jun 27, 2024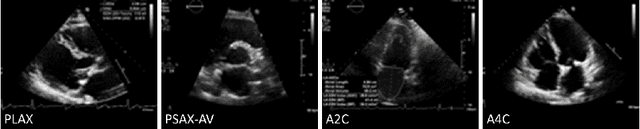
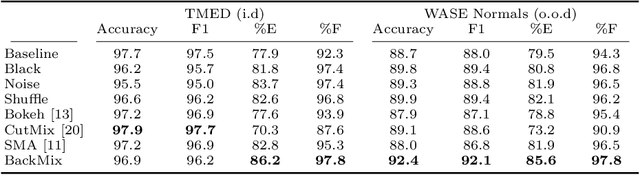
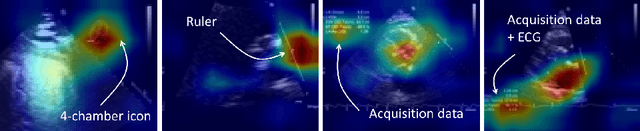
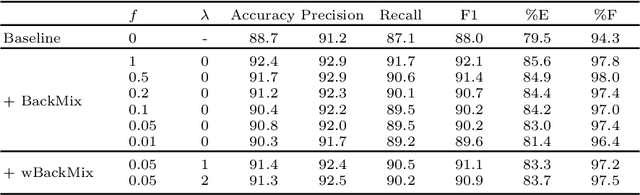
Abstract:Neural networks can learn spurious correlations that lead to the correct prediction in a validation set, but generalise poorly because the predictions are right for the wrong reason. This undesired learning of naive shortcuts (Clever Hans effect) can happen for example in echocardiogram view classification when background cues (e.g. metadata) are biased towards a class and the model learns to focus on those background features instead of on the image content. We propose a simple, yet effective random background augmentation method called BackMix, which samples random backgrounds from other examples in the training set. By enforcing the background to be uncorrelated with the outcome, the model learns to focus on the data within the ultrasound sector and becomes invariant to the regions outside this. We extend our method in a semi-supervised setting, finding that the positive effects of BackMix are maintained with as few as 5% of segmentation labels. A loss weighting mechanism, wBackMix, is also proposed to increase the contribution of the augmented examples. We validate our method on both in-distribution and out-of-distribution datasets, demonstrating significant improvements in classification accuracy, region focus and generalisability. Our source code is available at: https://github.com/kitbransby/BackMix
Contrastive Learning for View Classification of Echocardiograms
Aug 06, 2021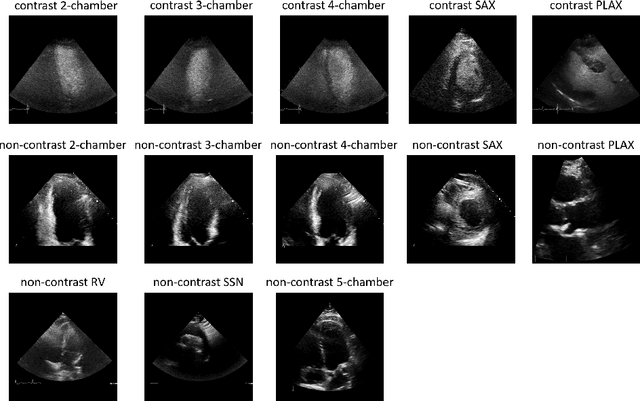
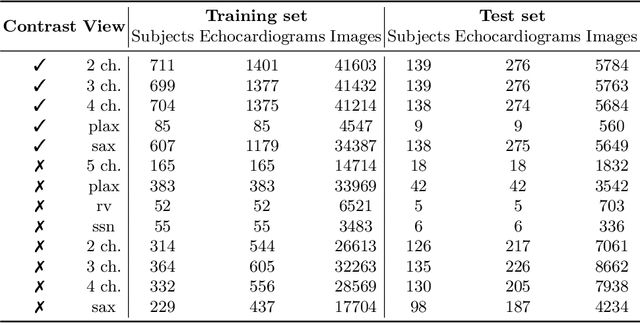
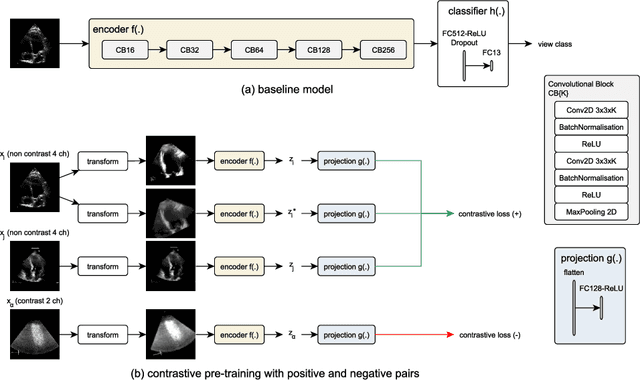
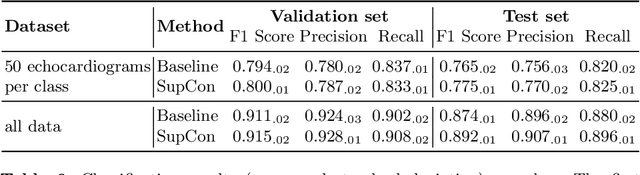
Abstract:Analysis of cardiac ultrasound images is commonly performed in routine clinical practice for quantification of cardiac function. Its increasing automation frequently employs deep learning networks that are trained to predict disease or detect image features. However, such models are extremely data-hungry and training requires labelling of many thousands of images by experienced clinicians. Here we propose the use of contrastive learning to mitigate the labelling bottleneck. We train view classification models for imbalanced cardiac ultrasound datasets and show improved performance for views/classes for which minimal labelled data is available. Compared to a naive baseline model, we achieve an improvement in F1 score of up to 26% in those views while maintaining state-of-the-art performance for the views with sufficiently many labelled training observations.
Max-Fusion U-Net for Multi-Modal Pathology Segmentation with Attention and Dynamic Resampling
Sep 05, 2020



Abstract:Automatic segmentation of multi-sequence (multi-modal) cardiac MR (CMR) images plays a significant role in diagnosis and management for a variety of cardiac diseases. However, the performance of relevant algorithms is significantly affected by the proper fusion of the multi-modal information. Furthermore, particular diseases, such as myocardial infarction, display irregular shapes on images and occupy small regions at random locations. These facts make pathology segmentation of multi-modal CMR images a challenging task. In this paper, we present the Max-Fusion U-Net that achieves improved pathology segmentation performance given aligned multi-modal images of LGE, T2-weighted, and bSSFP modalities. Specifically, modality-specific features are extracted by dedicated encoders. Then they are fused with the pixel-wise maximum operator. Together with the corresponding encoding features, these representations are propagated to decoding layers with U-Net skip-connections. Furthermore, a spatial-attention module is applied in the last decoding layer to encourage the network to focus on those small semantically meaningful pathological regions that trigger relatively high responses by the network neurons. We also use a simple image patch extraction strategy to dynamically resample training examples with varying spacial and batch sizes. With limited GPU memory, this strategy reduces the imbalance of classes and forces the model to focus on regions around the interested pathology. It further improves segmentation accuracy and reduces the mis-classification of pathology. We evaluate our methods using the Myocardial pathology segmentation (MyoPS) combining the multi-sequence CMR dataset which involves three modalities. Extensive experiments demonstrate the effectiveness of the proposed model which outperforms the related baselines.
* 13 pages, 7 figures, conference paper
Semi-supervised Pathology Segmentation with Disentangled Representations
Sep 05, 2020
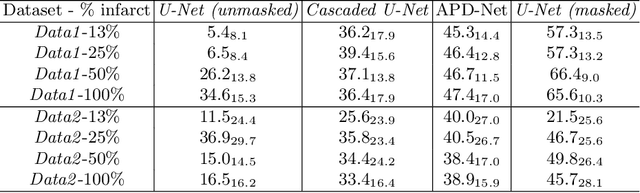
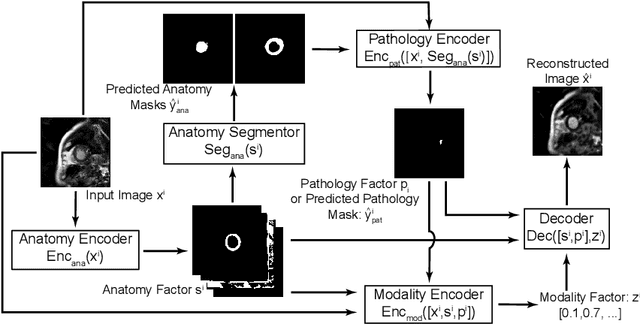
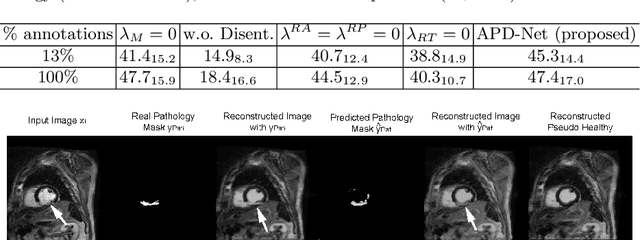
Abstract:Automated pathology segmentation remains a valuable diagnostic tool in clinical practice. However, collecting training data is challenging. Semi-supervised approaches by combining labelled and unlabelled data can offer a solution to data scarcity. An approach to semi-supervised learning relies on reconstruction objectives (as self-supervision objectives) that learns in a joint fashion suitable representations for the task. Here, we propose Anatomy-Pathology Disentanglement Network (APD-Net), a pathology segmentation model that attempts to learn jointly for the first time: disentanglement of anatomy, modality, and pathology. The model is trained in a semi-supervised fashion with new reconstruction losses directly aiming to improve pathology segmentation with limited annotations. In addition, a joint optimization strategy is proposed to fully take advantage of the available annotations. We evaluate our methods with two private cardiac infarction segmentation datasets with LGE-MRI scans. APD-Net can perform pathology segmentation with few annotations, maintain performance with different amounts of supervision, and outperform related deep learning methods.
* 12 Pages, 4 figures
Metrics for Exposing the Biases of Content-Style Disentanglement
Aug 31, 2020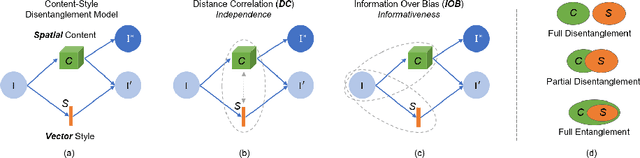
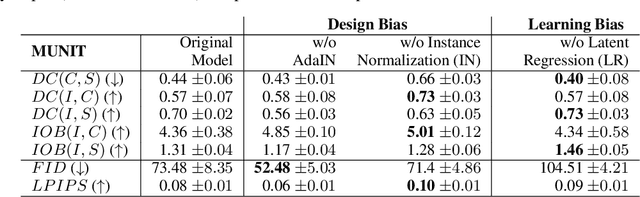
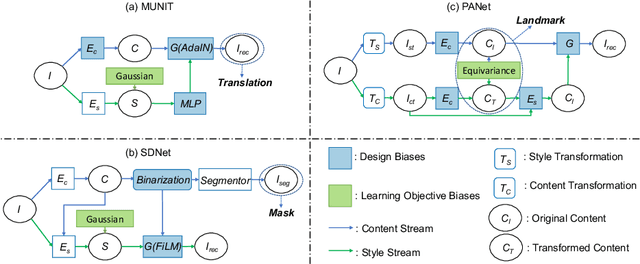

Abstract:Recent state-of-the-art semi- and un-supervised solutions for challenging computer vision tasks have used the idea of encoding image content into a spatial tensor and image appearance or "style" into a vector. These decomposed representations take advantage of equivariant properties of network design and improve performance in equivariant tasks, such as image-to-image translation. Most of these methods use the term "disentangled" for their representations and employ model design, learning objectives, and data biases to achieve good model performance. While considerable effort has been made to measure disentanglement in vector representations, currently, metrics that can characterize the degree of disentanglement between content (spatial) and style (vector) representations and the relation to task performance are lacking. In this paper, we propose metrics to measure how (un)correlated, biased, and informative the content and style representations are. In particular, we first identify key design choices and learning constraints on three popular models that employ content-style disentanglement and derive ablated versions. Then, we use our metrics to ascertain the role of each bias. Our experiments reveal a "sweet-spot" between disentanglement, task performance and latent space interpretability. The proposed metrics enable the design of better models and the selection of models that achieve the desired performance and disentanglement. Our metrics library is available at https://github.com/TsaftarisCollaboratory/CSDisentanglement_Metrics_Library.
Disentangled Representations for Domain-generalized Cardiac Segmentation
Aug 26, 2020


Abstract:Robust cardiac image segmentation is still an open challenge due to the inability of the existing methods to achieve satisfactory performance on unseen data of different domains. Since the acquisition and annotation of medical data are costly and time-consuming, recent work focuses on domain adaptation and generalization to bridge the gap between data from different populations and scanners. In this paper, we propose two data augmentation methods that focus on improving the domain adaptation and generalization abilities of state-to-the-art cardiac segmentation models. In particular, our "Resolution Augmentation" method generates more diverse data by rescaling images to different resolutions within a range spanning different scanner protocols. Subsequently, our "Factor-based Augmentation" method generates more diverse data by projecting the original samples onto disentangled latent spaces, and combining the learned anatomy and modality factors from different domains. Our extensive experiments demonstrate the importance of efficient adaptation between seen and unseen domains, as well as model generalization ability, to robust cardiac image segmentation.
Pseudo-healthy synthesis with pathology disentanglement and adversarial learning
May 05, 2020



Abstract:Pseudo-healthy synthesis is the task of creating a subject-specific `healthy' image from a pathological one. Such images can be helpful in tasks such as anomaly detection and understanding changes induced by pathology and disease. In this paper, we present a model that is encouraged to disentangle the information of pathology from what seems to be healthy. We disentangle what appears to be healthy and where disease is as a segmentation map, which are then recombined by a network to reconstruct the input disease image. We train our models adversarially using either paired or unpaired settings, where we pair disease images and maps when available. We quantitatively and subjectively, with a human study, evaluate the quality of pseudo-healthy images using several criteria. We show in a series of experiments, performed on ISLES, BraTS and Cam-CAN datasets, that our method is better than several baselines and methods from the literature. We also show that due to better training processes we could recover deformations, on surrounding tissue, caused by disease. Our implementation is publicly available at \url{https://tobeprovided.upon.acceptance}
Learning to synthesise the ageing brain without longitudinal data
Dec 12, 2019



Abstract:Brain ageing is a continuous process that is affected by many factors including neurodegenerative diseases. Understanding this process is of great value for both neuroscience research and clinical applications. However, revealing underlying mechanisms is challenging due to the lack of longitudinal data. In this paper, we propose a deep learning-based method that learns to simulate subject-specific brain ageing trajectories without relying on longitudinal data. Our method synthesises aged images using a network conditioned on two clinical variables: age as a continuous variable, and health state, i.e. status of Alzheimer's Disease (AD) for this work, as an ordinal variable. We adopt an adversarial loss to learn the joint distribution of brain appearance and clinical variables and define reconstruction losses that help preserve subject identity. To demonstrate our model, we compare with several approaches using two widely used datasets: Cam-CAN and ADNI. We use ground-truth longitudinal data from ADNI to evaluate the quality of synthesised images. A pre-trained age predictor, which estimates the apparent age of a brain image, is used to assess age accuracy. In addition, we show that we can train the model on Cam-CAN data and evaluate on the longitudinal data from ADNI, indicating the generalisation power of our approach. Both qualitative and quantitative results show that our method can progressively simulate the ageing process by synthesising realistic brain images. The code will be made publicly available at: https://github.com/xiat0616/BrainAgeing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge