Angela Mumith
Left Ventricle Contouring of Apical Three-Chamber Views on 2D Echocardiography
Jul 13, 2022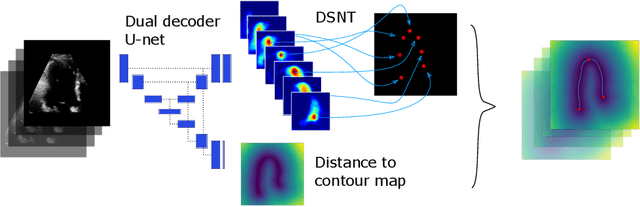
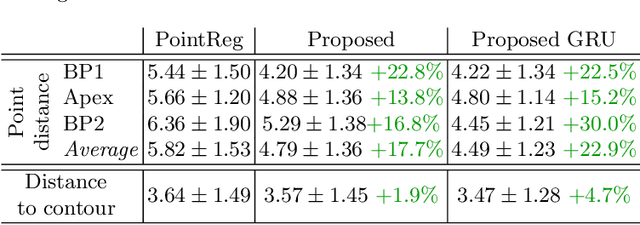
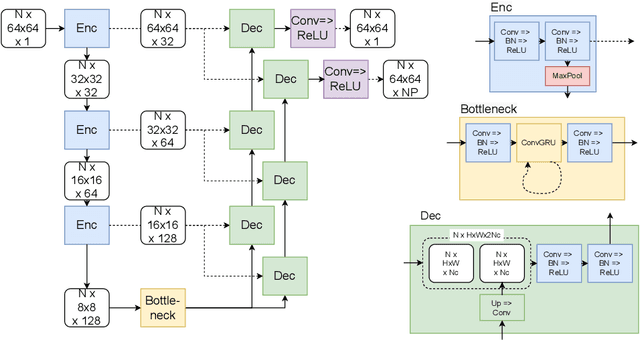
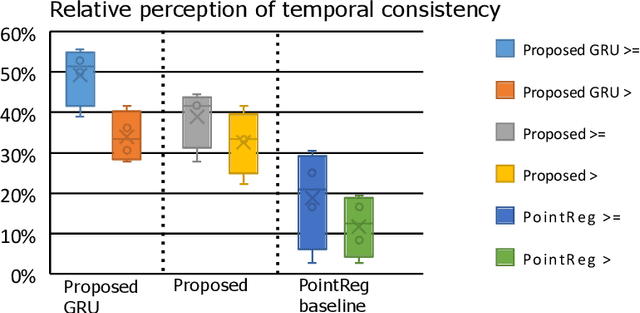
Abstract:We propose a new method to automatically contour the left ventricle on 2D echocardiographic images. Unlike most existing segmentation methods, which are based on predicting segmentation masks, we focus at predicting the endocardial contour and the key landmark points within this contour (basal points and apex). This provides a representation that is closer to how experts perform manual annotations and hence produce results that are physiologically more plausible. Our proposed method uses a two-headed network based on the U-Net architecture. One head predicts the 7 contour points, and the other head predicts a distance map to the contour. This approach was compared to the U-Net and to a point based approach, achieving performance gains of up to 30\% in terms of landmark localisation (<4.5mm) and distance to the ground truth contour (<3.5mm).
Contrastive Learning for View Classification of Echocardiograms
Aug 06, 2021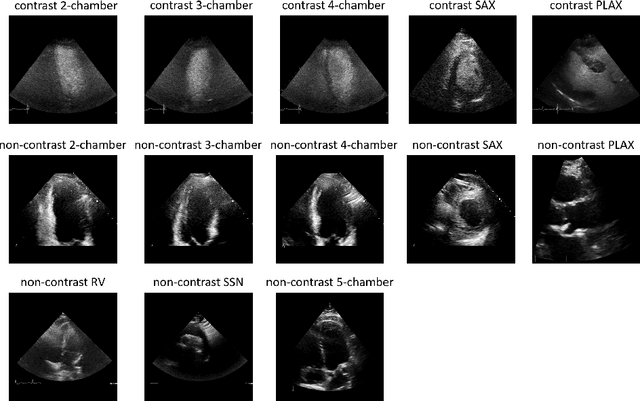
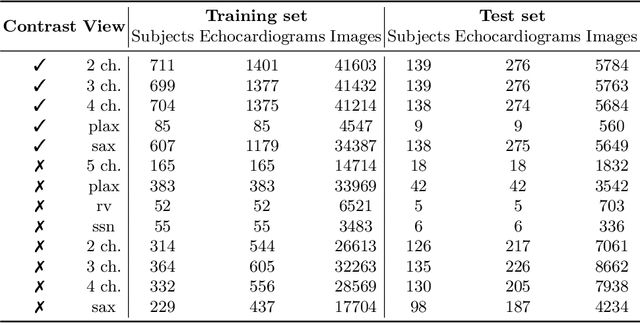
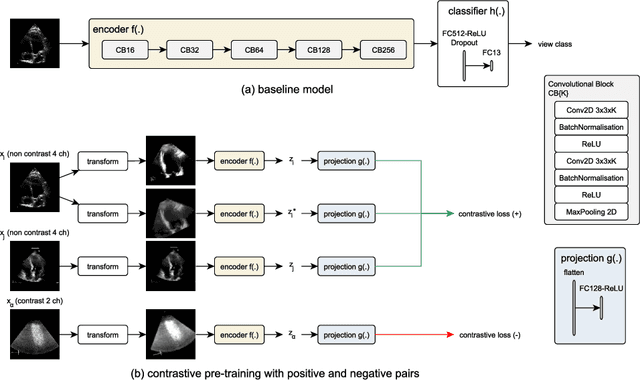
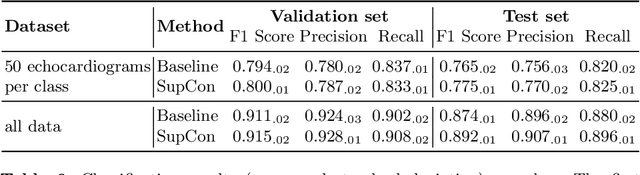
Abstract:Analysis of cardiac ultrasound images is commonly performed in routine clinical practice for quantification of cardiac function. Its increasing automation frequently employs deep learning networks that are trained to predict disease or detect image features. However, such models are extremely data-hungry and training requires labelling of many thousands of images by experienced clinicians. Here we propose the use of contrastive learning to mitigate the labelling bottleneck. We train view classification models for imbalanced cardiac ultrasound datasets and show improved performance for views/classes for which minimal labelled data is available. Compared to a naive baseline model, we achieve an improvement in F1 score of up to 26% in those views while maintaining state-of-the-art performance for the views with sufficiently many labelled training observations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge