Mihaela Porumb
Left Ventricle Contouring of Apical Three-Chamber Views on 2D Echocardiography
Jul 13, 2022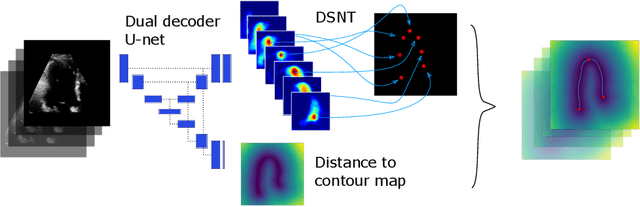
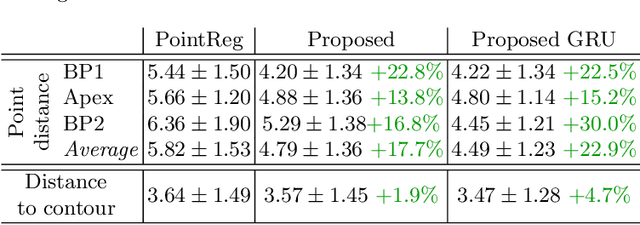
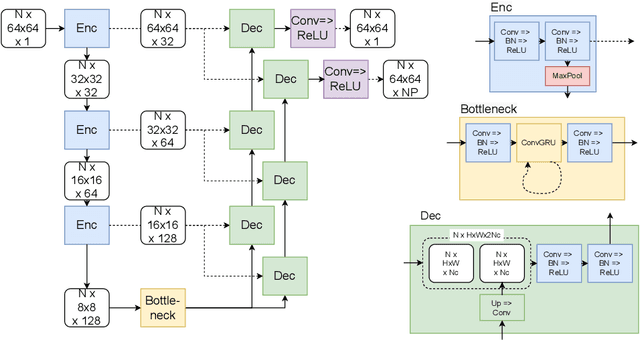
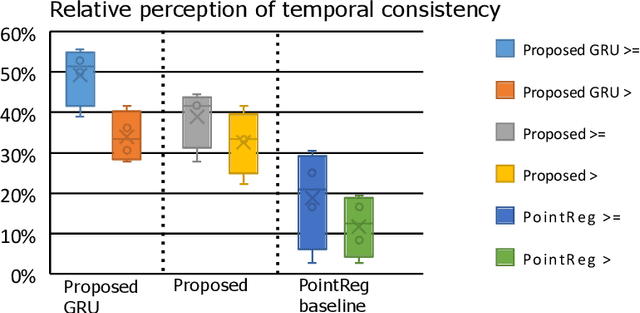
Abstract:We propose a new method to automatically contour the left ventricle on 2D echocardiographic images. Unlike most existing segmentation methods, which are based on predicting segmentation masks, we focus at predicting the endocardial contour and the key landmark points within this contour (basal points and apex). This provides a representation that is closer to how experts perform manual annotations and hence produce results that are physiologically more plausible. Our proposed method uses a two-headed network based on the U-Net architecture. One head predicts the 7 contour points, and the other head predicts a distance map to the contour. This approach was compared to the U-Net and to a point based approach, achieving performance gains of up to 30\% in terms of landmark localisation (<4.5mm) and distance to the ground truth contour (<3.5mm).
CRISP - Reliable Uncertainty Estimation for Medical Image Segmentation
Jun 15, 2022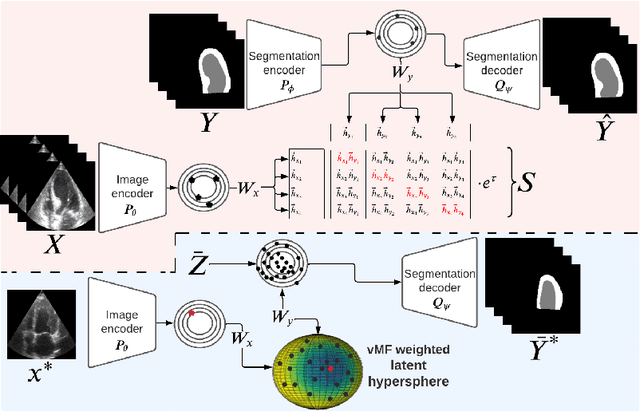
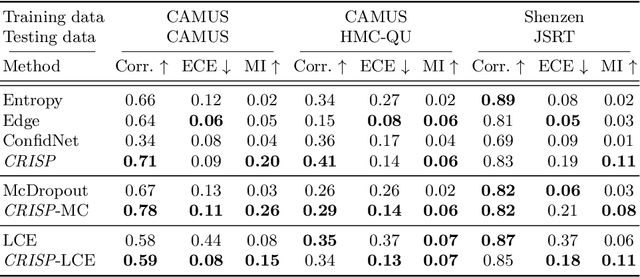
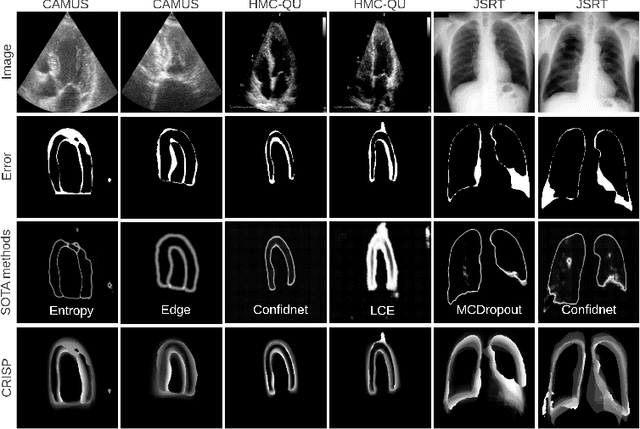
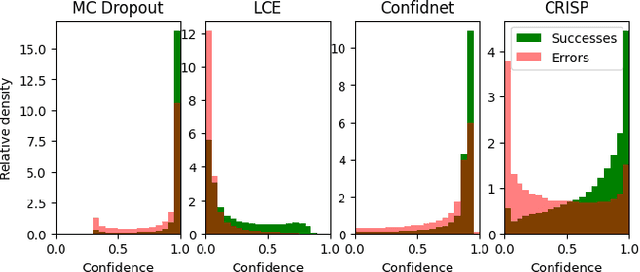
Abstract:Accurate uncertainty estimation is a critical need for the medical imaging community. A variety of methods have been proposed, all direct extensions of classification uncertainty estimations techniques. The independent pixel-wise uncertainty estimates, often based on the probabilistic interpretation of neural networks, do not take into account anatomical prior knowledge and consequently provide sub-optimal results to many segmentation tasks. For this reason, we propose CRISP a ContRastive Image Segmentation for uncertainty Prediction method. At its core, CRISP implements a contrastive method to learn a joint latent space which encodes a distribution of valid segmentations and their corresponding images. We use this joint latent space to compare predictions to thousands of latent vectors and provide anatomically consistent uncertainty maps. Comprehensive studies performed on four medical image databases involving different modalities and organs underlines the superiority of our method compared to state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge