Olivier Bernard
Reinforcement Learning for Unsupervised Domain Adaptation in Spatio-Temporal Echocardiography Segmentation
Oct 16, 2025Abstract:Domain adaptation methods aim to bridge the gap between datasets by enabling knowledge transfer across domains, reducing the need for additional expert annotations. However, many approaches struggle with reliability in the target domain, an issue particularly critical in medical image segmentation, where accuracy and anatomical validity are essential. This challenge is further exacerbated in spatio-temporal data, where the lack of temporal consistency can significantly degrade segmentation quality, and particularly in echocardiography, where the presence of artifacts and noise can further hinder segmentation performance. To address these issues, we present RL4Seg3D, an unsupervised domain adaptation framework for 2D + time echocardiography segmentation. RL4Seg3D integrates novel reward functions and a fusion scheme to enhance key landmark precision in its segmentations while processing full-sized input videos. By leveraging reinforcement learning for image segmentation, our approach improves accuracy, anatomical validity, and temporal consistency while also providing, as a beneficial side effect, a robust uncertainty estimator, which can be used at test time to further enhance segmentation performance. We demonstrate the effectiveness of our framework on over 30,000 echocardiographic videos, showing that it outperforms standard domain adaptation techniques without the need for any labels on the target domain. Code is available at https://github.com/arnaudjudge/RL4Seg3D.
DAFTED: Decoupled Asymmetric Fusion of Tabular and Echocardiographic Data for Cardiac Hypertension Diagnosis
Sep 19, 2025Abstract:Multimodal data fusion is a key approach for enhancing diagnosis in medical applications. We propose an asymmetric fusion strategy starting from a primary modality and integrating secondary modalities by disentangling shared and modality-specific information. Validated on a dataset of 239 patients with echocardiographic time series and tabular records, our model outperforms existing methods, achieving an AUC over 90%. This improvement marks a crucial benchmark for clinical use.
Generation of realistic cardiac ultrasound sequences with ground truth motion and speckle decorrelation
Sep 05, 2025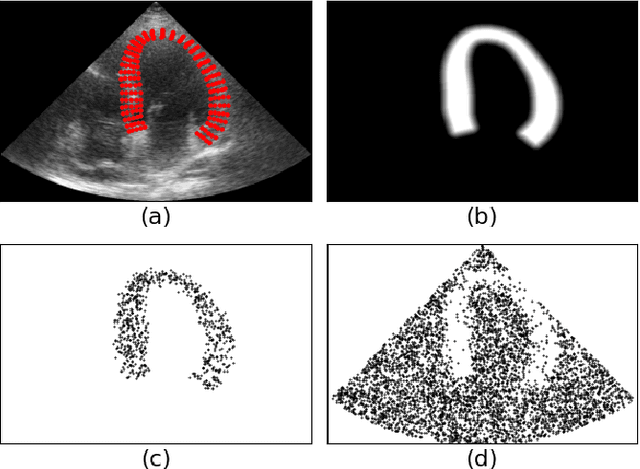
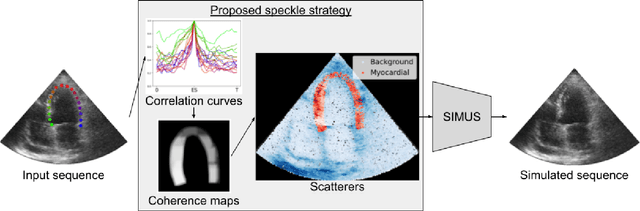
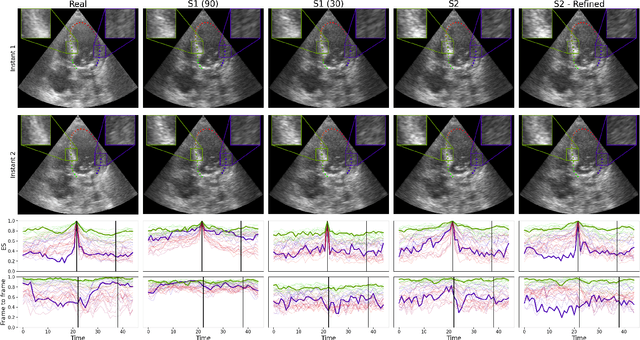
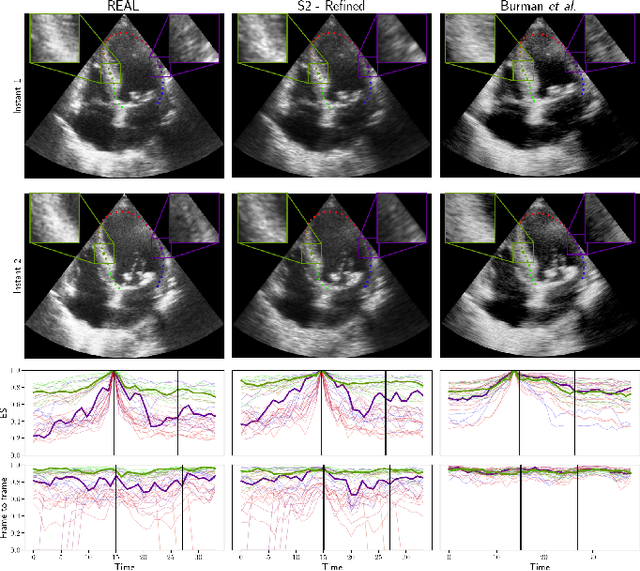
Abstract:Simulated ultrasound image sequences are key for training and validating machine learning algorithms for left ventricular strain estimation. Several simulation pipelines have been proposed to generate sequences with corresponding ground truth motion, but they suffer from limited realism as they do not consider speckle decorrelation. In this work, we address this limitation by proposing an improved simulation framework that explicitly accounts for speckle decorrelation. Our method builds on an existing ultrasound simulation pipeline by incorporating a dynamic model of speckle variation. Starting from real ultrasound sequences and myocardial segmentations, we generate meshes that guide image formation. Instead of applying a fixed ratio of myocardial and background scatterers, we introduce a coherence map that adapts locally over time. This map is derived from correlation values measured directly from the real ultrasound data, ensuring that simulated sequences capture the characteristic temporal changes observed in practice. We evaluated the realism of our approach using ultrasound data from 98 patients in the CAMUS database. Performance was assessed by comparing correlation curves from real and simulated images. The proposed method achieved lower mean absolute error compared to the baseline pipeline, indicating that it more faithfully reproduces the decorrelation behavior seen in clinical data.
Uncertainty Propagation for Echocardiography Clinical Metric Estimation via Contour Sampling
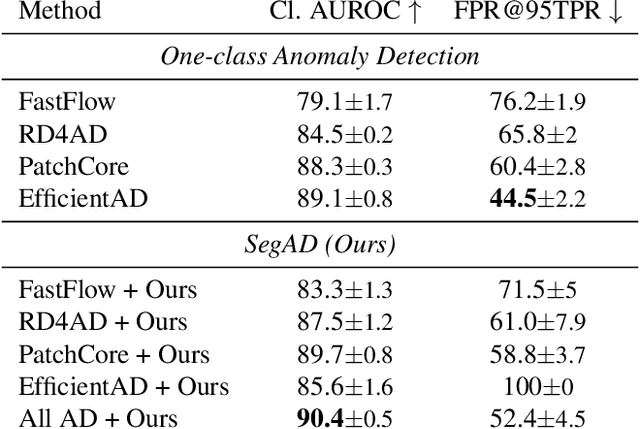
Feb 18, 2025Abstract:Echocardiography plays a fundamental role in the extraction of important clinical parameters (e.g. left ventricular volume and ejection fraction) required to determine the presence and severity of heart-related conditions. When deploying automated techniques for computing these parameters, uncertainty estimation is crucial for assessing their utility. Since clinical parameters are usually derived from segmentation maps, there is no clear path for converting pixel-wise uncertainty values into uncertainty estimates in the downstream clinical metric calculation. In this work, we propose a novel uncertainty estimation method based on contouring rather than segmentation. Our method explicitly predicts contour location uncertainty from which contour samples can be drawn. Finally, the sampled contours can be used to propagate uncertainty to clinical metrics. Our proposed method not only provides accurate uncertainty estimations for the task of contouring but also for the downstream clinical metrics on two cardiac ultrasound datasets. Code is available at: https://github.com/ThierryJudge/contouring-uncertainty.
Domain Adaptation of Echocardiography Segmentation Via Reinforcement Learning
Jun 25, 2024



Abstract:Performance of deep learning segmentation models is significantly challenged in its transferability across different medical imaging domains, particularly when aiming to adapt these models to a target domain with insufficient annotated data for effective fine-tuning. While existing domain adaptation (DA) methods propose strategies to alleviate this problem, these methods do not explicitly incorporate human-verified segmentation priors, compromising the potential of a model to produce anatomically plausible segmentations. We introduce RL4Seg, an innovative reinforcement learning framework that reduces the need to otherwise incorporate large expertly annotated datasets in the target domain, and eliminates the need for lengthy manual human review. Using a target dataset of 10,000 unannotated 2D echocardiographic images, RL4Seg not only outperforms existing state-of-the-art DA methods in accuracy but also achieves 99% anatomical validity on a subset of 220 expert-validated subjects from the target domain. Furthermore, our framework's reward network offers uncertainty estimates comparable with dedicated state-of-the-art uncertainty methods, demonstrating the utility and effectiveness of RL4Seg in overcoming domain adaptation challenges in medical image segmentation.
Supervised Anomaly Detection for Complex Industrial Images
May 08, 2024
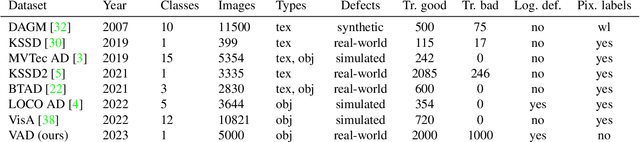

Abstract:Automating visual inspection in industrial production lines is essential for increasing product quality across various industries. Anomaly detection (AD) methods serve as robust tools for this purpose. However, existing public datasets primarily consist of images without anomalies, limiting the practical application of AD methods in production settings. To address this challenge, we present (1) the Valeo Anomaly Dataset (VAD), a novel real-world industrial dataset comprising 5000 images, including 2000 instances of challenging real defects across more than 20 subclasses. Acknowledging that traditional AD methods struggle with this dataset, we introduce (2) Segmentation-based Anomaly Detector (SegAD). First, SegAD leverages anomaly maps as well as segmentation maps to compute local statistics. Next, SegAD uses these statistics and an optional supervised classifier score as input features for a Boosted Random Forest (BRF) classifier, yielding the final anomaly score. Our SegAD achieves state-of-the-art performance on both VAD (+2.1% AUROC) and the VisA dataset (+0.4% AUROC). The code and the models are publicly available.
Physics-Guided Neural Networks for Intraventricular Vector Flow Mapping
Mar 19, 2024



Abstract:Intraventricular vector flow mapping (iVFM) seeks to enhance and quantify color Doppler in cardiac imaging. In this study, we propose novel alternatives to the traditional iVFM optimization scheme by utilizing physics-informed neural networks (PINNs) and a physics-guided nnU-Net-based supervised approach. Through rigorous evaluation on simulated color Doppler images derived from a patient-specific computational fluid dynamics model and in vivo Doppler acquisitions, both approaches demonstrate comparable reconstruction performance to the original iVFM algorithm. The efficiency of PINNs is boosted through dual-stage optimization and pre-optimized weights. On the other hand, the nnU-Net method excels in generalizability and real time capabilities. Notably, nnU-Net shows superior robustness on sparse and truncated Doppler data while maintaining independence from explicit boundary conditions. Overall, our results highlight the effectiveness of these methods in reconstructing intraventricular vector blood flow. The study also suggests potential applications of PINNs in ultrafast color Doppler imaging and the incorporation of fluid dynamics equations to derive biomarkers for cardiovascular diseases based on blood flow.
Fusing Echocardiography Images and Medical Records for Continuous Patient Stratification
Jan 15, 2024Abstract:Deep learning now enables automatic and robust extraction of cardiac function descriptors from echocardiographic sequences, such as ejection fraction or strain. These descriptors provide fine-grained information that physicians consider, in conjunction with more global variables from the clinical record, to assess patients' condition. Drawing on novel transformer models applied to tabular data (e.g., variables from electronic health records), we propose a method that considers all descriptors extracted from medical records and echocardiograms to learn the representation of a difficult-to-characterize cardiovascular pathology, namely hypertension. Our method first projects each variable into its own representation space using modality-specific approaches. These standardized representations of multimodal data are then fed to a transformer encoder, which learns to merge them into a comprehensive representation of the patient through a pretext task of predicting a clinical rating. This pretext task is formulated as an ordinal classification to enforce a pathological continuum in the representation space. We observe the major trends along this continuum for a cohort of 239 hypertensive patients to describe, with unprecedented gradation, the effect of hypertension on a number of cardiac function descriptors. Our analysis shows that i) pretrained weights from a foundation model allow to reach good performance (83% accuracy) even with limited data (less than 200 training samples), ii) trends across the population are reproducible between trainings, and iii) for descriptors whose interactions with hypertension are well documented, patterns are consistent with prior physiological knowledge.
Phase Unwrapping of Color Doppler Echocardiography using Deep Learning
Jul 05, 2023



Abstract:Color Doppler echocardiography is a widely used non-invasive imaging modality that provides real-time information about the intracardiac blood flow. In an apical long-axis view of the left ventricle, color Doppler is subject to phase wrapping, or aliasing, especially during cardiac filling and ejection. When setting up quantitative methods based on color Doppler, it is necessary to correct this wrapping artifact. We developed an unfolded primal-dual network to unwrap (dealias) color Doppler echocardiographic images and compared its effectiveness against two state-of-the-art segmentation approaches based on nnU-Net and transformer models. We trained and evaluated the performance of each method on an in-house dataset and found that the nnU-Net-based method provided the best dealiased results, followed by the primal-dual approach and the transformer-based technique. Noteworthy, the primal-dual network, which had significantly fewer trainable parameters, performed competitively with respect to the other two methods, demonstrating the high potential of deep unfolding methods. Our results suggest that deep learning-based methods can effectively remove aliasing artifacts in color Doppler echocardiographic images, outperforming DeAN, a state-of-the-art semi-automatic technique. Overall, our results show that deep learning-based methods have the potential to effectively preprocess color Doppler images for downstream quantitative analysis.
Ultrafast Cardiac Imaging Using Deep Learning For Speckle-Tracking Echocardiography
Jun 25, 2023
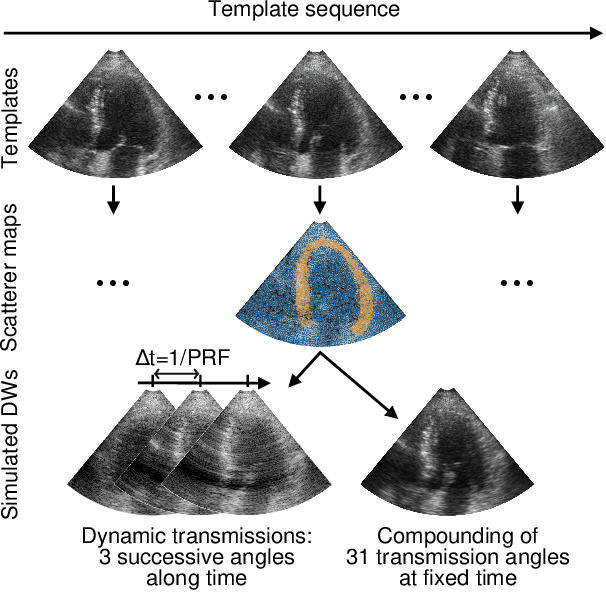
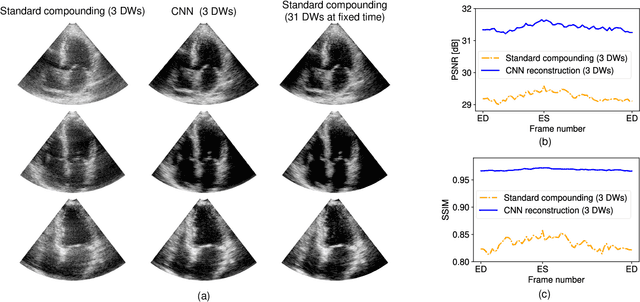
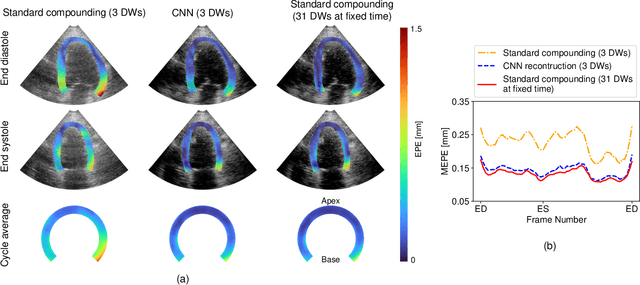
Abstract:High-quality ultrafast ultrasound imaging is based on coherent compounding from multiple transmissions of plane waves (PW) or diverging waves (DW). However, compounding results in reduced frame rate, as well as destructive interferences from high-velocity tissue motion if motion compensation (MoCo) is not considered. While many studies have recently shown the interest of deep learning for the reconstruction of high-quality static images from PW or DW, its ability to achieve such performance while maintaining the capability of tracking cardiac motion has yet to be assessed. In this paper, we addressed such issue by deploying a complex-weighted convolutional neural network (CNN) for image reconstruction and a state-of-the-art speckle tracking method. The evaluation of this approach was first performed by designing an adapted simulation framework, which provides specific reference data, i.e. high quality, motion artifact-free cardiac images. The obtained results showed that, while using only three DWs as input, the CNN-based approach yielded an image quality and a motion accuracy equivalent to those obtained by compounding 31 DWs free of motion artifacts. The performance was then further evaluated on non-simulated, experimental in vitro data, using a spinning disk phantom. This experiment demonstrated that our approach yielded high-quality image reconstruction and motion estimation, under a large range of velocities and outperforms a state-of-the-art MoCo-based approach at high velocities. Our method was finally assessed on in vivo datasets and showed consistent improvement in image quality and motion estimation compared to standard compounding. This demonstrates the feasibility and effectiveness of deep learning reconstruction for ultrafast speckle-tracking echocardiography.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge