Jean Provost
CREATIS
Functional Assessment of Cerebral Capillaries using Single Capillary Reporters in Ultrasound Localization Microscopy
Jul 11, 2024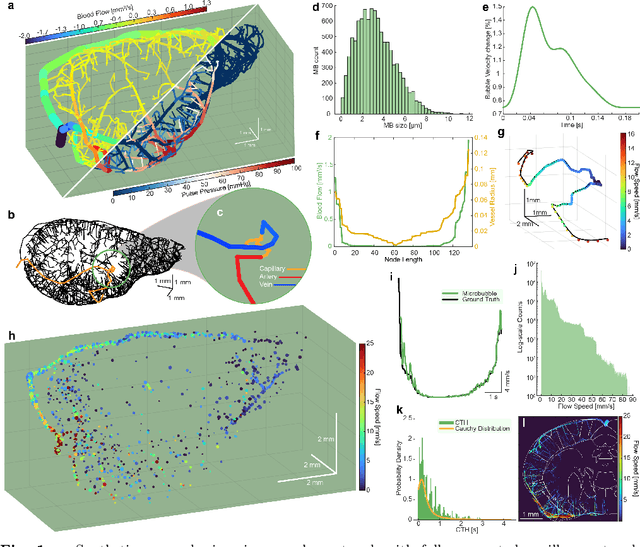



Abstract:The brain's microvascular cerebral capillary network plays a vital role in maintaining neuronal health, yet capillary dynamics are still not well understood due to limitations in existing imaging techniques. Here, we present Single Capillary Reporters (SCaRe) for transcranial Ultrasound Localization Microscopy (ULM), a novel approach enabling non-invasive, whole-brain mapping of single capillaries and estimates of their transit-time as a neurovascular biomarker. We accomplish this first through computational Monte Carlo and ultrasound simulations of microbubbles flowing through a fully-connected capillary network. We unveil distinct capillary flow behaviors which informs methodological changes to ULM acquisitions to better capture capillaries in vivo. Subsequently, applying SCaRe-ULM in vivo, we achieve unprecedented visualization of single capillary tracks across brain regions, analysis of layer-specific capillary heterogeneous transit times (CHT), and characterization of whole microbubble trajectories from arterioles to venules. Lastly, we evaluate capillary biomarkers using injected lipopolysaccharide to induce systemic neuroinflammation and track the increase in SCaRe-ULM CHT, demonstrating the capability to detect subtle capillary functional changes. SCaRe-ULM represents a significant advance in studying microvascular dynamics, offering novel avenues for investigating capillary patterns in neurological disorders and potential diagnostic applications.
Towards Transcranial 3D Ultrasound Localization Microscopy of the Nonhuman Primate Brain
Apr 04, 2024



Abstract:Hemodynamic changes occur in stroke and neurodegenerative diseases. Developing imaging techniques allowing the in vivo visualization and quantification of cerebral blood flow would help better understand the underlying mechanism of those cerebrovascular diseases. 3D ultrasound localization microscopy (ULM) is a novel technology that can map the microvasculature of the brain at large depth and has been mainly used until now in rodents. Here, we demonstrated the feasibility of 3D ULM of the nonhuman primate (NHP) brain with a single 256-channels programmable ultrasound scanner. We achieved a highly resolved vascular map of the macaque brain at large depth in presence of craniotomy and durectomy using an 8-MHz multiplexed matrix probe. We were able to distinguish vessels as small as 26.9 {\mu}m. We also demonstrated that transcranial imaging of the macaque brain at similar depth was feasible using a 3-MHz probe and achieved a resolution of 60.4 {\mu}m. This work paves the way to clinical application of 3D ULM.
Boosting Cardiac Color Doppler Frame Rates with Deep Learning
Mar 28, 2024Abstract:Color Doppler echocardiography enables visualization of blood flow within the heart. However, the limited frame rate impedes the quantitative assessment of blood velocity throughout the cardiac cycle, thereby compromising a comprehensive analysis of ventricular filling. Concurrently, deep learning is demonstrating promising outcomes in post-processing of echocardiographic data for various applications. This work explores the use of deep learning models for intracardiac Doppler velocity estimation from a reduced number of filtered I/Q signals. We used a supervised learning approach by simulating patient-based cardiac color Doppler acquisitions and proposed data augmentation strategies to enlarge the training dataset. We implemented architectures based on convolutional neural networks. In particular, we focused on comparing the U-Net model and the recent ConvNeXt models, alongside assessing real-valued versus complex-valued representations. We found that both models outperformed the state-of-the-art autocorrelator method, effectively mitigating aliasing and noise. We did not observe significant differences between the use of real and complex data. Finally, we validated the models on in vitro and in vivo experiments. All models produced quantitatively comparable results to the baseline and were more robust to noise. ConvNeXt emerged as the sole model to achieve high-quality results on in vivo aliased samples. These results demonstrate the interest of supervised deep learning methods for Doppler velocity estimation from a reduced number of acquisitions.
Pruning Sparse Tensor Neural Networks Enables Deep Learning for 3D Ultrasound Localization Microscopy
Feb 14, 2024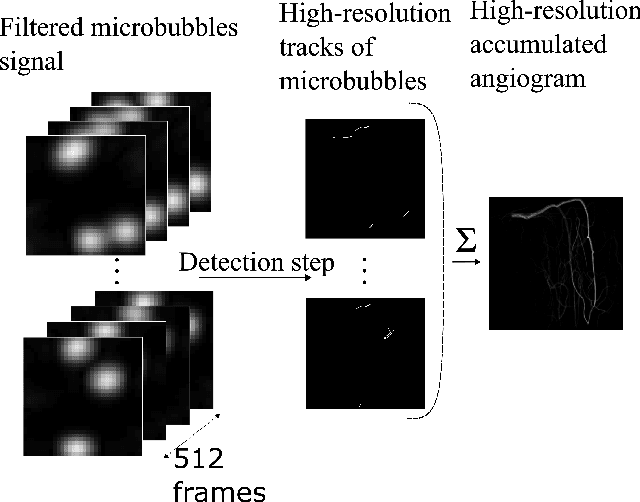
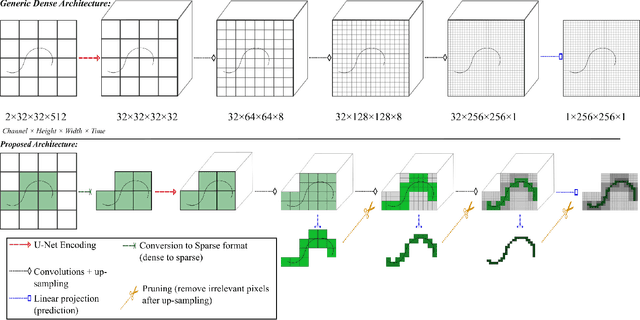
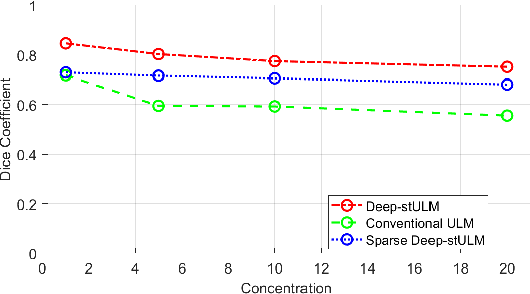
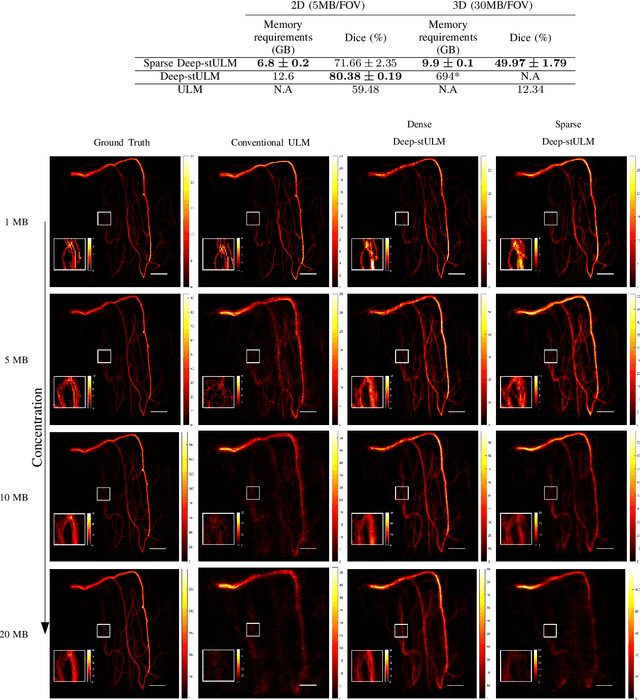
Abstract:Ultrasound Localization Microscopy (ULM) is a non-invasive technique that allows for the imaging of micro-vessels in vivo, at depth and with a resolution on the order of ten microns. ULM is based on the sub-resolution localization of individual microbubbles injected in the bloodstream. Mapping the whole angioarchitecture requires the accumulation of microbubbles trajectories from thousands of frames, typically acquired over a few minutes. ULM acquisition times can be reduced by increasing the microbubble concentration, but requires more advanced algorithms to detect them individually. Several deep learning approaches have been proposed for this task, but they remain limited to 2D imaging, in part due to the associated large memory requirements. Herein, we propose to use sparse tensor neural networks to reduce memory usage in 2D and to improve the scaling of the memory requirement for the extension of deep learning architecture to 3D. We study several approaches to efficiently convert ultrasound data into a sparse format and study the impact of the associated loss of information. When applied in 2D, the sparse formulation reduces the memory requirements by a factor 2 at the cost of a small reduction of performance when compared against dense networks. In 3D, the proposed approach reduces memory requirements by two order of magnitude while largely outperforming conventional ULM in high concentration settings. We show that Sparse Tensor Neural Networks in 3D ULM allow for the same benefits as dense deep learning based method in 2D ULM i.e. the use of higher concentration in silico and reduced acquisition time.
Inverse Problem Based on a Sparse Representation of Contrast-enhanced Ultrasound Data for in vivo Transcranial Imaging
Jan 18, 2024Abstract:Transcranial ultrasound imaging is currently limited by attenuation and aberration induced by the skull. First used in contrast-enhanced ultrasound (CEUS), highly echoic microbubbles allowed for the development of novel imaging modalities such as ultrasound localization microscopy (ULM). Herein, we develop an inverse problem approach to aberration correction (IPAC) that leverages the sparsity of microbubble signals. We propose to use the \textit{a priori} knowledge of the medium based upon microbubble localization and wave propagation to build a forward model to link the measured signals directly to the aberration function. A standard least-squares inversion is then used to retrieve the aberration function. We first validated IPAC on simulated data of a vascular network using plane wave as well as divergent wave emissions. We then evaluated the reproducibility of IPAC \textit{in vivo} in 5 mouse brains. We showed that aberration correction improved the contrast of CEUS images by 4.6 dB. For ULM images, IPAC yielded sharper vessels, reduced vessel duplications, and improved the resolution from 21.1 $\mu$m to 18.3 $\mu$m. Aberration correction also improved hemodynamic quantification for velocity magnitude and flow direction.
A Deep Learning Framework for Spatiotemporal Ultrasound Localization Microscopy
Oct 12, 2023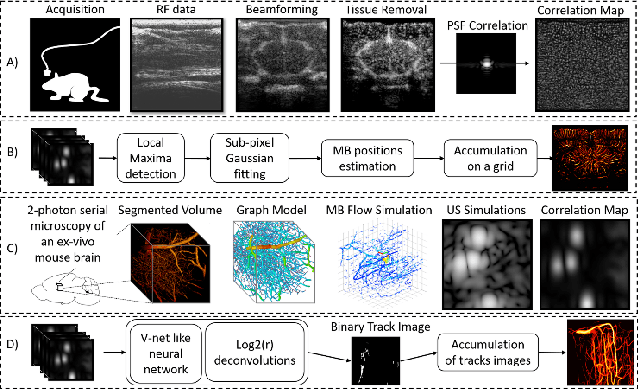
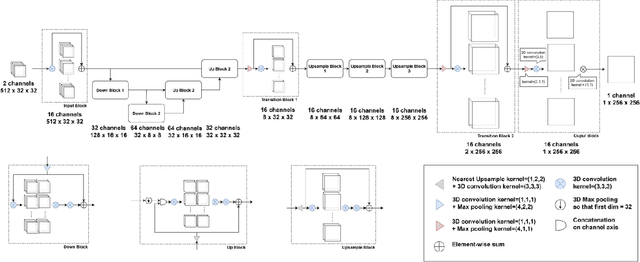
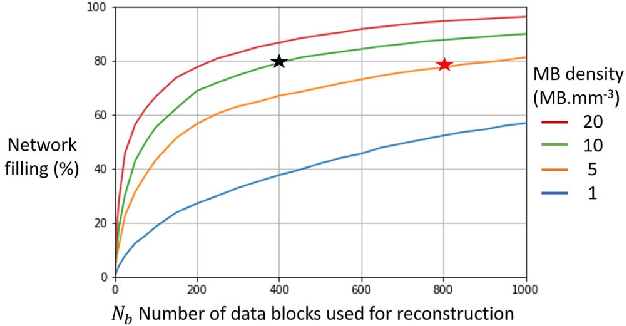
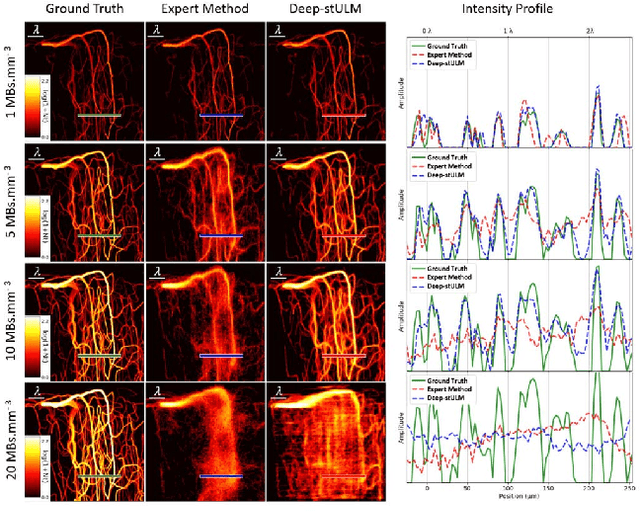
Abstract:Ultrasound Localization Microscopy can resolve the microvascular bed down to a few micrometers. To achieve such performance microbubble contrast agents must perfuse the entire microvascular network. Microbubbles are then located individually and tracked over time to sample individual vessels, typically over hundreds of thousands of images. To overcome the fundamental limit of diffraction and achieve a dense reconstruction of the network, low microbubble concentrations must be used, which lead to acquisitions lasting several minutes. Conventional processing pipelines are currently unable to deal with interference from multiple nearby microbubbles, further reducing achievable concentrations. This work overcomes this problem by proposing a Deep Learning approach to recover dense vascular networks from ultrasound acquisitions with high microbubble concentrations. A realistic mouse brain microvascular network, segmented from 2-photon microscopy, was used to train a three-dimensional convolutional neural network based on a V-net architecture. Ultrasound data sets from multiple microbubbles flowing through the microvascular network were simulated and used as ground truth to train the 3D CNN to track microbubbles. The 3D-CNN approach was validated in silico using a subset of the data and in vivo on a rat brain acquisition. In silico, the CNN reconstructed vascular networks with higher precision (81%) than a conventional ULM framework (70%). In vivo, the CNN could resolve micro vessels as small as 10 $\mu$m with an increase in resolution when compared against a conventional approach.
* Copyright 2021 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works
A Tracking prior to Localization workflow for Ultrasound Localization Microscopy
Aug 04, 2023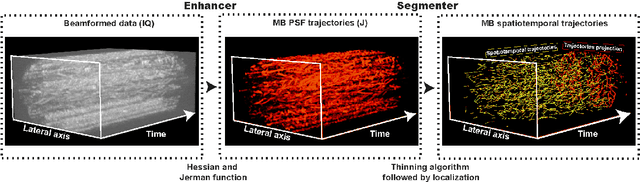
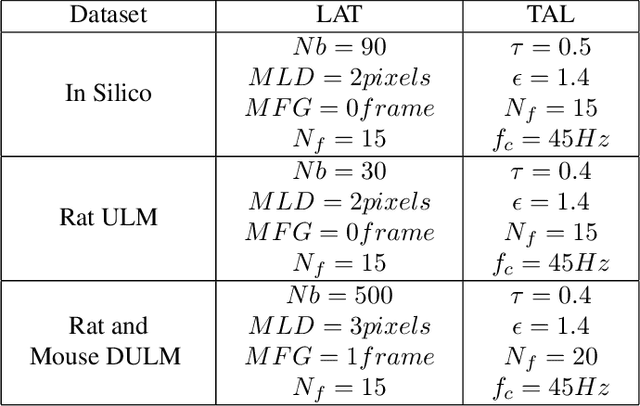
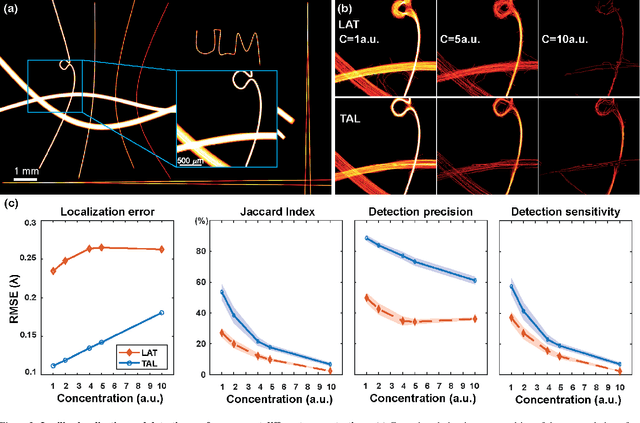
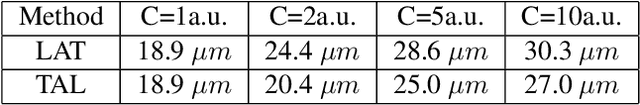
Abstract:Ultrasound Localization Microscopy (ULM) has proven effective in resolving microvascular structures and local mean velocities at sub-diffraction-limited scales, offering high-resolution imaging capabilities. Dynamic ULM (DULM) enables the creation of angiography or velocity movies throughout cardiac cycles. Currently, these techniques rely on a Localization-and-Tracking (LAT) workflow consisting in detecting microbubbles (MB) in the frames before pairing them to generate tracks. While conventional LAT methods perform well at low concentrations, they suffer from longer acquisition times and degraded localization and tracking accuracy at higher concentrations, leading to biased angiogram reconstruction and velocity estimation. In this study, we propose a novel approach to address these challenges by reversing the current workflow. The proposed method, Tracking-and-Localization (TAL), relies on first tracking the MB and then performing localization. Through comprehensive benchmarking using both in silico and in vivo experiments and employing various metrics to quantify ULM angiography and velocity maps, we demonstrate that the TAL method consistently outperforms the reference LAT workflow. Moreover, when applied to DULM, TAL successfully extracts velocity variations along the cardiac cycle with improved repeatability. The findings of this work highlight the effectiveness of the TAL approach in overcoming the limitations of conventional LAT methods, providing enhanced ULM angiography and velocity imaging.
Ultrafast Cardiac Imaging Using Deep Learning For Speckle-Tracking Echocardiography
Jun 25, 2023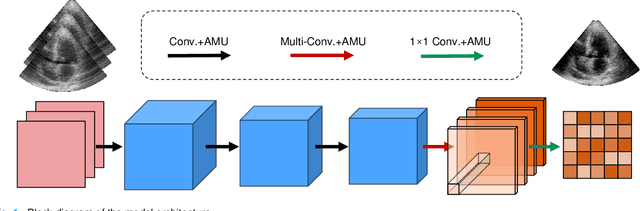
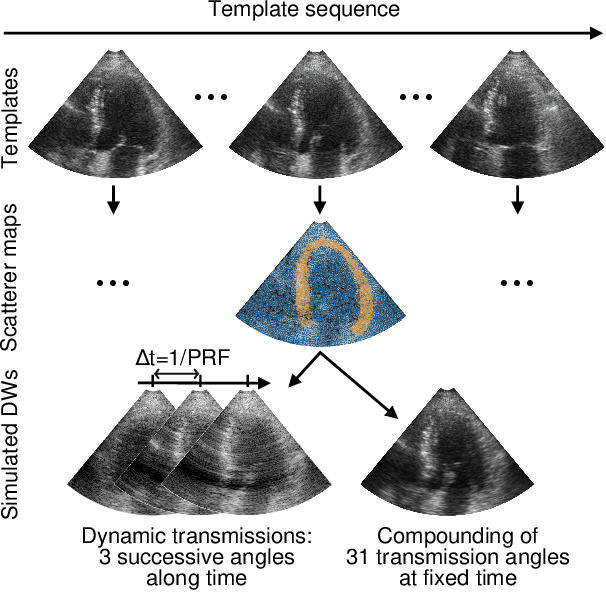
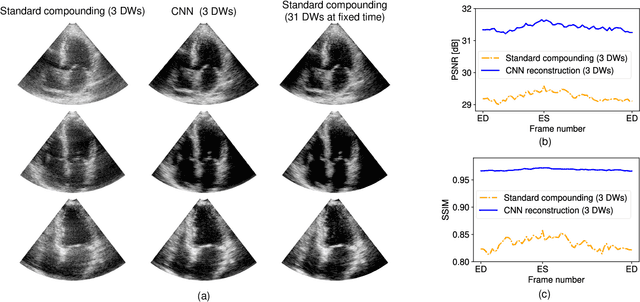
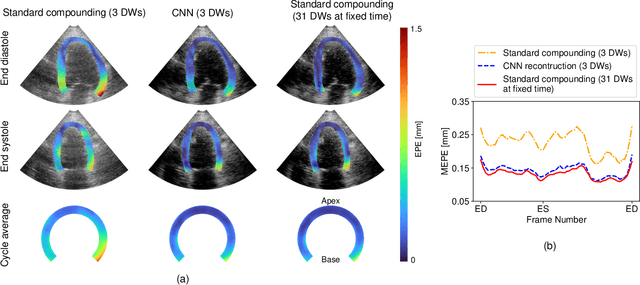
Abstract:High-quality ultrafast ultrasound imaging is based on coherent compounding from multiple transmissions of plane waves (PW) or diverging waves (DW). However, compounding results in reduced frame rate, as well as destructive interferences from high-velocity tissue motion if motion compensation (MoCo) is not considered. While many studies have recently shown the interest of deep learning for the reconstruction of high-quality static images from PW or DW, its ability to achieve such performance while maintaining the capability of tracking cardiac motion has yet to be assessed. In this paper, we addressed such issue by deploying a complex-weighted convolutional neural network (CNN) for image reconstruction and a state-of-the-art speckle tracking method. The evaluation of this approach was first performed by designing an adapted simulation framework, which provides specific reference data, i.e. high quality, motion artifact-free cardiac images. The obtained results showed that, while using only three DWs as input, the CNN-based approach yielded an image quality and a motion accuracy equivalent to those obtained by compounding 31 DWs free of motion artifacts. The performance was then further evaluated on non-simulated, experimental in vitro data, using a spinning disk phantom. This experiment demonstrated that our approach yielded high-quality image reconstruction and motion estimation, under a large range of velocities and outperforms a state-of-the-art MoCo-based approach at high velocities. Our method was finally assessed on in vivo datasets and showed consistent improvement in image quality and motion estimation compared to standard compounding. This demonstrates the feasibility and effectiveness of deep learning reconstruction for ultrafast speckle-tracking echocardiography.
Phase Aberration Correction for in vivo Ultrasound Localization Microscopy Using a Spatiotemporal Complex-Valued Neural Network
Sep 23, 2022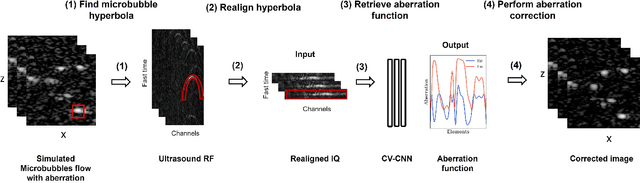
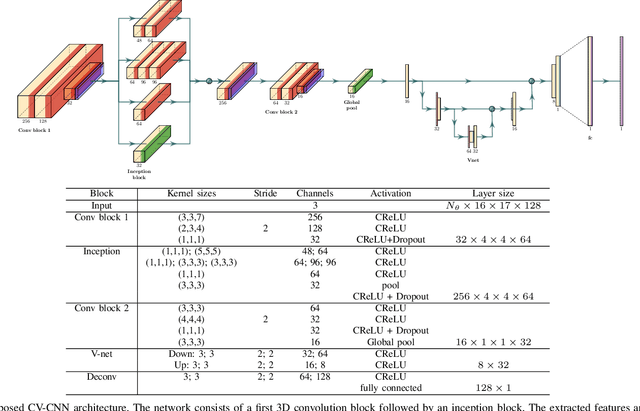
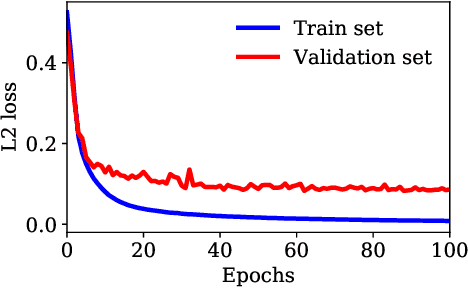
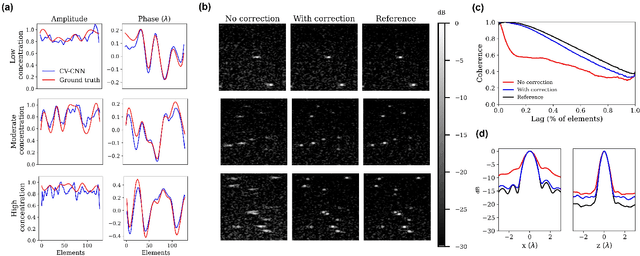
Abstract:Ultrasound Localization Microscopy (ULM) can map microvessels at a resolution of a few micrometers ({\mu}m). Transcranial ULM remains challenging in presence of aberrations caused by the skull, which lead to localization errors. Herein, we propose a deep learning approach based on recently introduced complex-valued convolutional neural networks (CV-CNNs) to retrieve the aberration function, which can then be used to form enhanced images using standard delay-and-sum beamforming. Complex-valued convolutional networks were selected as they can apply time delays through multiplication with in-phase quadrature input data. Predicting the aberration function rather than corrected images also confers enhanced explainability to the network. In addition, 3D spatiotemporal convolutions were used for the network to leverage entire microbubble tracks. For training and validation, we used an anatomically and hemodynamically realistic mouse brain microvascular network model to simulate the flow of microbubbles in presence of aberration. We then confirmed the capability of our network to generalize to transcranial in vivo data in the mouse brain (n=2). Qualitatively, vascular reconstructions using a pixel-wise predicted aberration function included additional and sharper vessels. The spatial resolution was evaluated by using the Fourier ring correlation (FRC). After correction, we measured a resolution of 16.7 {\mu}m in vivo, representing an improvement of up to 27.5 %. This work leads to different applications for complex-valued convolutions in biomedical imaging and strategies to perform transcranial ULM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge