Brice Rauby
Polytechnique Montréal, Mila - Quebec Artificial Intelligence Institute
GitChameleon: Evaluating AI Code Generation Against Python Library Version Incompatibilities
Jul 16, 2025
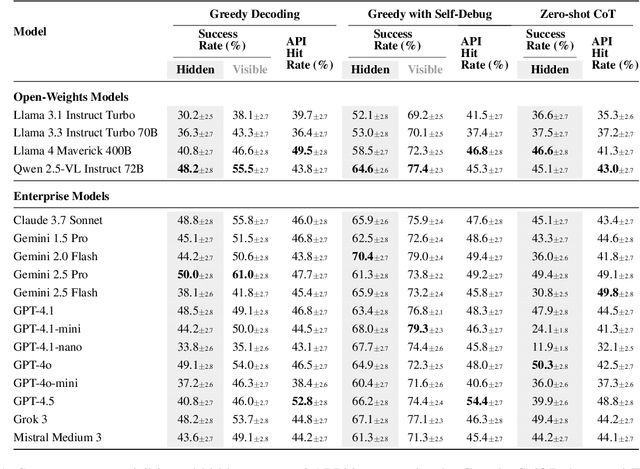
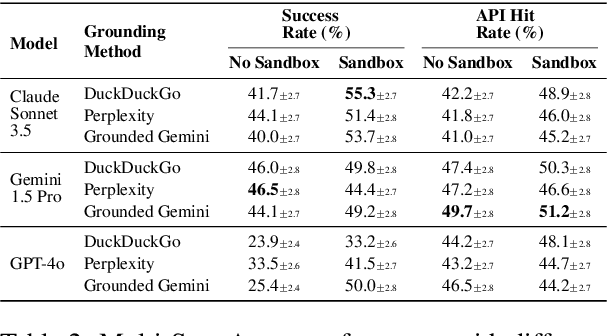
Abstract:The rapid evolution of software libraries poses a considerable hurdle for code generation, necessitating continuous adaptation to frequent version updates while preserving backward compatibility. While existing code evolution benchmarks provide valuable insights, they typically lack execution-based evaluation for generating code compliant with specific library versions. To address this, we introduce GitChameleon, a novel, meticulously curated dataset comprising 328 Python code completion problems, each conditioned on specific library versions and accompanied by executable unit tests. GitChameleon rigorously evaluates the capacity of contemporary large language models (LLMs), LLM-powered agents, code assistants, and RAG systems to perform version-conditioned code generation that demonstrates functional accuracy through execution. Our extensive evaluations indicate that state-of-the-art systems encounter significant challenges with this task; enterprise models achieving baseline success rates in the 48-51\% range, underscoring the intricacy of the problem. By offering an execution-based benchmark emphasizing the dynamic nature of code libraries, GitChameleon enables a clearer understanding of this challenge and helps guide the development of more adaptable and dependable AI code generation methods. We make the dataset and evaluation code publicly available at https://github.com/mrcabbage972/GitChameleonBenchmark.
Pruning Sparse Tensor Neural Networks Enables Deep Learning for 3D Ultrasound Localization Microscopy
Feb 14, 2024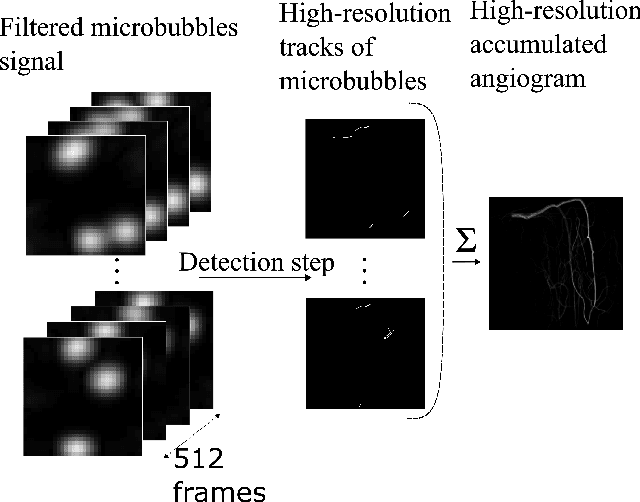
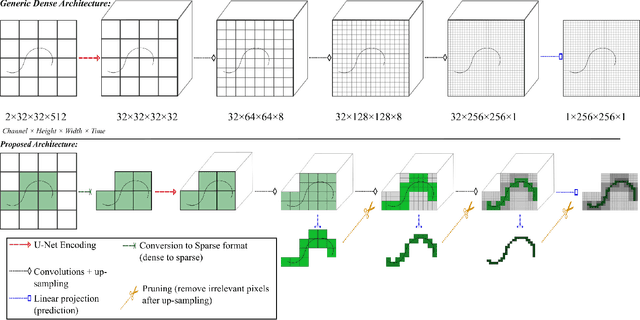
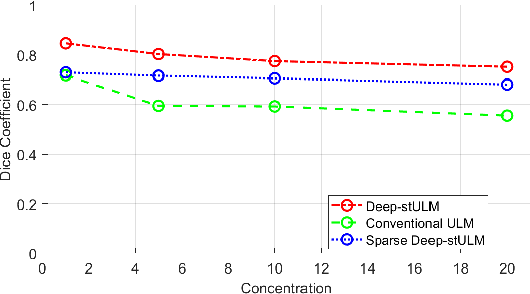
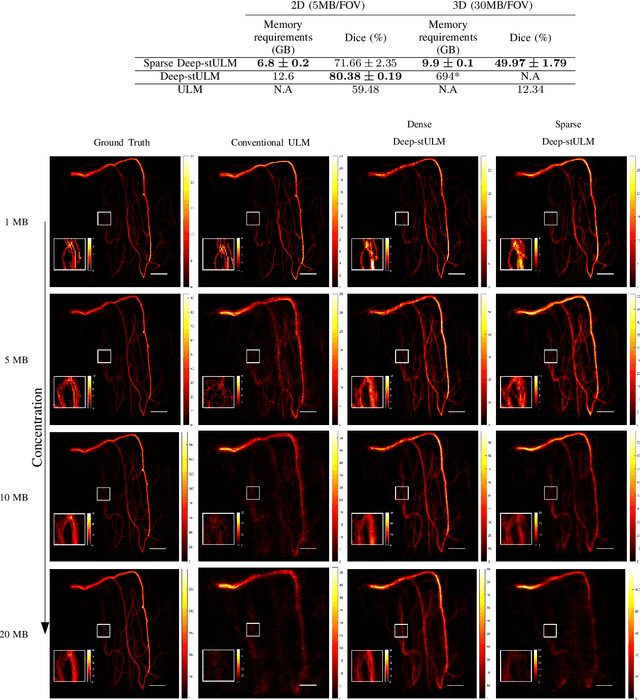
Abstract:Ultrasound Localization Microscopy (ULM) is a non-invasive technique that allows for the imaging of micro-vessels in vivo, at depth and with a resolution on the order of ten microns. ULM is based on the sub-resolution localization of individual microbubbles injected in the bloodstream. Mapping the whole angioarchitecture requires the accumulation of microbubbles trajectories from thousands of frames, typically acquired over a few minutes. ULM acquisition times can be reduced by increasing the microbubble concentration, but requires more advanced algorithms to detect them individually. Several deep learning approaches have been proposed for this task, but they remain limited to 2D imaging, in part due to the associated large memory requirements. Herein, we propose to use sparse tensor neural networks to reduce memory usage in 2D and to improve the scaling of the memory requirement for the extension of deep learning architecture to 3D. We study several approaches to efficiently convert ultrasound data into a sparse format and study the impact of the associated loss of information. When applied in 2D, the sparse formulation reduces the memory requirements by a factor 2 at the cost of a small reduction of performance when compared against dense networks. In 3D, the proposed approach reduces memory requirements by two order of magnitude while largely outperforming conventional ULM in high concentration settings. We show that Sparse Tensor Neural Networks in 3D ULM allow for the same benefits as dense deep learning based method in 2D ULM i.e. the use of higher concentration in silico and reduced acquisition time.
A Tracking prior to Localization workflow for Ultrasound Localization Microscopy
Aug 04, 2023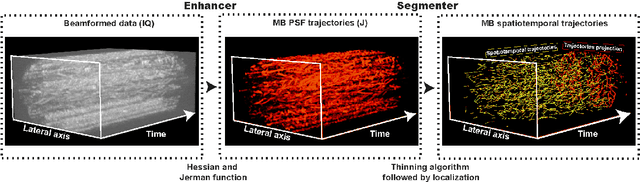
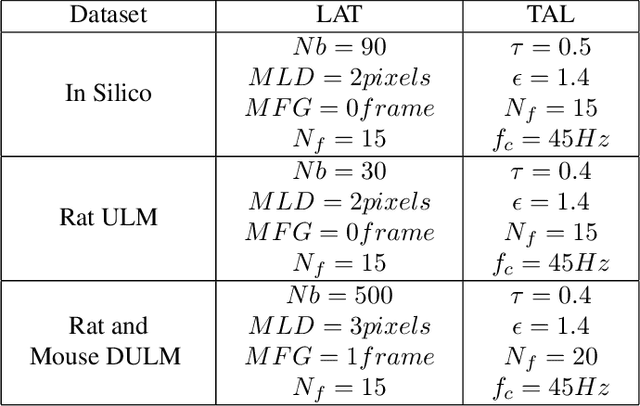
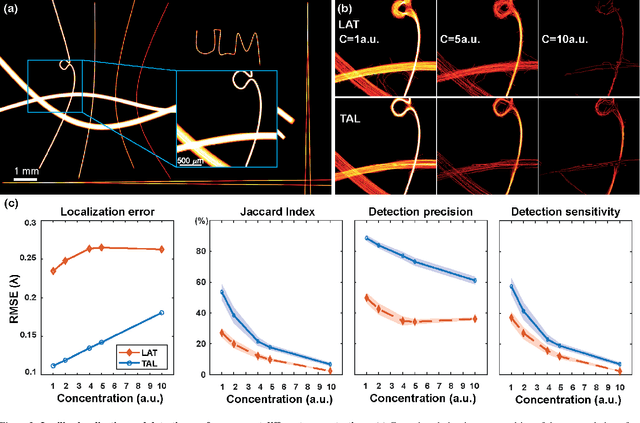
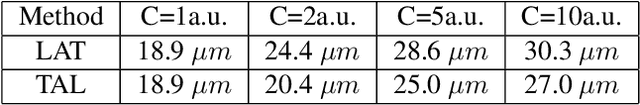
Abstract:Ultrasound Localization Microscopy (ULM) has proven effective in resolving microvascular structures and local mean velocities at sub-diffraction-limited scales, offering high-resolution imaging capabilities. Dynamic ULM (DULM) enables the creation of angiography or velocity movies throughout cardiac cycles. Currently, these techniques rely on a Localization-and-Tracking (LAT) workflow consisting in detecting microbubbles (MB) in the frames before pairing them to generate tracks. While conventional LAT methods perform well at low concentrations, they suffer from longer acquisition times and degraded localization and tracking accuracy at higher concentrations, leading to biased angiogram reconstruction and velocity estimation. In this study, we propose a novel approach to address these challenges by reversing the current workflow. The proposed method, Tracking-and-Localization (TAL), relies on first tracking the MB and then performing localization. Through comprehensive benchmarking using both in silico and in vivo experiments and employing various metrics to quantify ULM angiography and velocity maps, we demonstrate that the TAL method consistently outperforms the reference LAT workflow. Moreover, when applied to DULM, TAL successfully extracts velocity variations along the cardiac cycle with improved repeatability. The findings of this work highlight the effectiveness of the TAL approach in overcoming the limitations of conventional LAT methods, providing enhanced ULM angiography and velocity imaging.
Phase Aberration Correction for in vivo Ultrasound Localization Microscopy Using a Spatiotemporal Complex-Valued Neural Network
Sep 23, 2022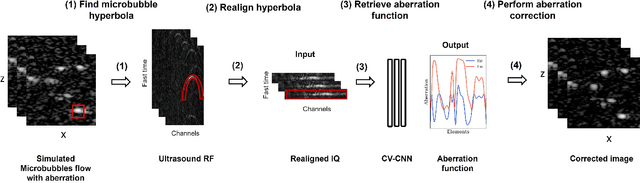
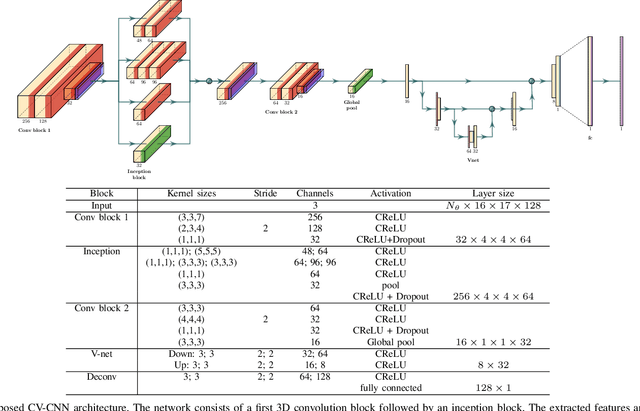
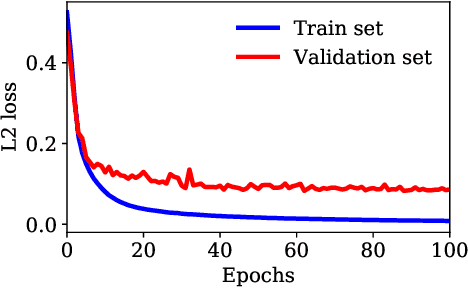
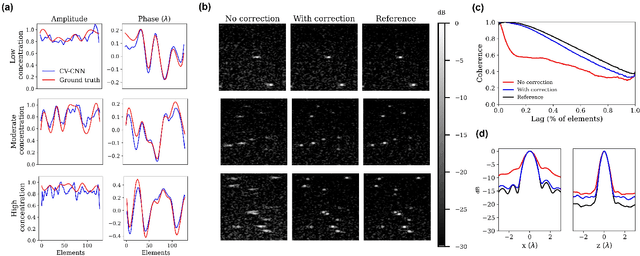
Abstract:Ultrasound Localization Microscopy (ULM) can map microvessels at a resolution of a few micrometers ({\mu}m). Transcranial ULM remains challenging in presence of aberrations caused by the skull, which lead to localization errors. Herein, we propose a deep learning approach based on recently introduced complex-valued convolutional neural networks (CV-CNNs) to retrieve the aberration function, which can then be used to form enhanced images using standard delay-and-sum beamforming. Complex-valued convolutional networks were selected as they can apply time delays through multiplication with in-phase quadrature input data. Predicting the aberration function rather than corrected images also confers enhanced explainability to the network. In addition, 3D spatiotemporal convolutions were used for the network to leverage entire microbubble tracks. For training and validation, we used an anatomically and hemodynamically realistic mouse brain microvascular network model to simulate the flow of microbubbles in presence of aberration. We then confirmed the capability of our network to generalize to transcranial in vivo data in the mouse brain (n=2). Qualitatively, vascular reconstructions using a pixel-wise predicted aberration function included additional and sharper vessels. The spatial resolution was evaluated by using the Fourier ring correlation (FRC). After correction, we measured a resolution of 16.7 {\mu}m in vivo, representing an improvement of up to 27.5 %. This work leads to different applications for complex-valued convolutions in biomedical imaging and strategies to perform transcranial ULM.
Geo-Spatiotemporal Features and Shape-Based Prior Knowledge for Fine-grained Imbalanced Data Classification
Mar 21, 2021
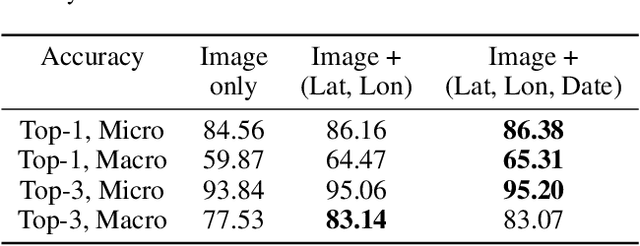

Abstract:Fine-grained classification aims at distinguishing between items with similar global perception and patterns, but that differ by minute details. Our primary challenges come from both small inter-class variations and large intra-class variations. In this article, we propose to combine several innovations to improve fine-grained classification within the use-case of wildlife, which is of practical interest for experts. We utilize geo-spatiotemporal data to enrich the picture information and further improve the performance. We also investigate state-of-the-art methods for handling the imbalanced data issue.
* Copyright by the authors. All rights reserved to authors only. Correspondence to: ckantor (at) stanford [dot] edu
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge