Kit Mills Bransby
BackMix: Mitigating Shortcut Learning in Echocardiography with Minimal Supervision
Jun 27, 2024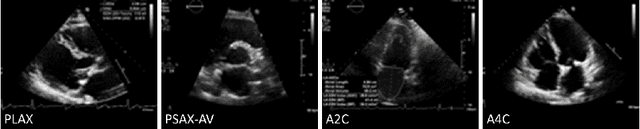
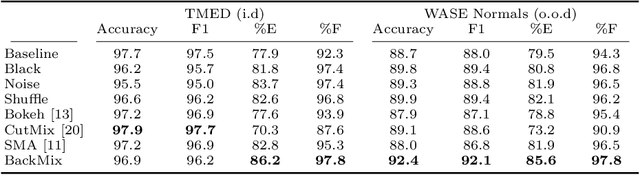
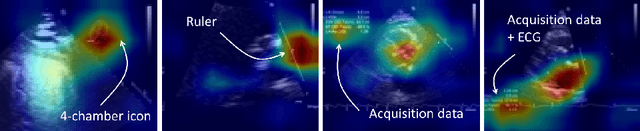
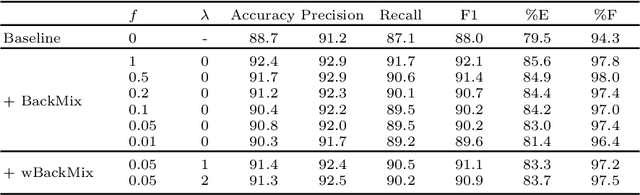
Abstract:Neural networks can learn spurious correlations that lead to the correct prediction in a validation set, but generalise poorly because the predictions are right for the wrong reason. This undesired learning of naive shortcuts (Clever Hans effect) can happen for example in echocardiogram view classification when background cues (e.g. metadata) are biased towards a class and the model learns to focus on those background features instead of on the image content. We propose a simple, yet effective random background augmentation method called BackMix, which samples random backgrounds from other examples in the training set. By enforcing the background to be uncorrelated with the outcome, the model learns to focus on the data within the ultrasound sector and becomes invariant to the regions outside this. We extend our method in a semi-supervised setting, finding that the positive effects of BackMix are maintained with as few as 5% of segmentation labels. A loss weighting mechanism, wBackMix, is also proposed to increase the contribution of the augmented examples. We validate our method on both in-distribution and out-of-distribution datasets, demonstrating significant improvements in classification accuracy, region focus and generalisability. Our source code is available at: https://github.com/kitbransby/BackMix
Joint Dense-Point Representation for Contour-Aware Graph Segmentation
Jun 21, 2023Abstract:We present a novel methodology that combines graph and dense segmentation techniques by jointly learning both point and pixel contour representations, thereby leveraging the benefits of each approach. This addresses deficiencies in typical graph segmentation methods where misaligned objectives restrict the network from learning discriminative vertex and contour features. Our joint learning strategy allows for rich and diverse semantic features to be encoded, while alleviating common contour stability issues in dense-based approaches, where pixel-level objectives can lead to anatomically implausible topologies. In addition, we identify scenarios where correct predictions that fall on the contour boundary are penalised and address this with a novel hybrid contour distance loss. Our approach is validated on several Chest X-ray datasets, demonstrating clear improvements in segmentation stability and accuracy against a variety of dense- and point-based methods. Our source code is freely available at: www.github.com/kitbransby/Joint_Graph_Segmentation
3D Coronary Vessel Reconstruction from Bi-Plane Angiography using Graph Convolutional Networks
Feb 28, 2023
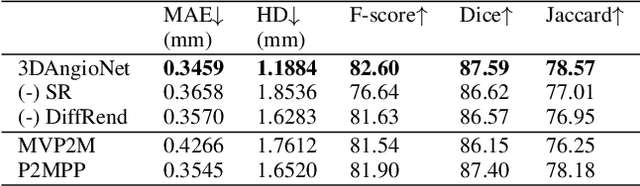


Abstract:X-ray coronary angiography (XCA) is used to assess coronary artery disease and provides valuable information on lesion morphology and severity. However, XCA images are 2D and therefore limit visualisation of the vessel. 3D reconstruction of coronary vessels is possible using multiple views, however lumen border detection in current software is performed manually resulting in limited reproducibility and slow processing time. In this study we propose 3DAngioNet, a novel deep learning (DL) system that enables rapid 3D vessel mesh reconstruction using 2D XCA images from two views. Our approach learns a coarse mesh template using an EfficientB3-UNet segmentation network and projection geometries, and deforms it using a graph convolutional network. 3DAngioNet outperforms similar automated reconstruction methods, offers improved efficiency, and enables modelling of bifurcated vessels. The approach was validated using state-of-the-art software verified by skilled cardiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge