Qianni Zhang
Joint Neural Networks for One-shot Object Recognition and Detection
Aug 01, 2024



Abstract:This paper presents a novel joint neural networks approach to address the challenging one-shot object recognition and detection tasks. Inspired by Siamese neural networks and state-of-art multi-box detection approaches, the joint neural networks are able to perform object recognition and detection for categories that remain unseen during the training process. Following the one-shot object recognition/detection constraints, the training and testing datasets do not contain overlapped classes, in other words, all the test classes remain unseen during training. The joint networks architecture is able to effectively compare pairs of images via stacked convolutional layers of the query and target inputs, recognising patterns of the same input query category without relying on previous training around this category. The proposed approach achieves 61.41% accuracy for one-shot object recognition on the MiniImageNet dataset and 47.1% mAP for one-shot object detection when trained on the COCO dataset and tested using the Pascal VOC dataset. Code available at https://github.com/cjvargasc/JNN recog and https://github.com/cjvargasc/JNN detection/
STS MICCAI 2023 Challenge: Grand challenge on 2D and 3D semi-supervised tooth segmentation
Jul 18, 2024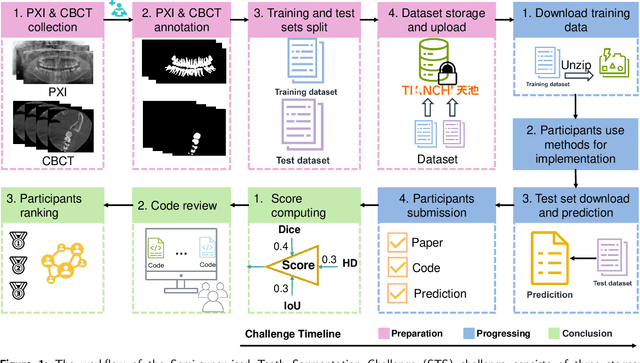
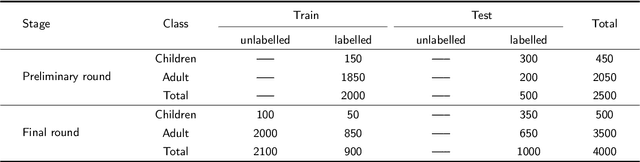
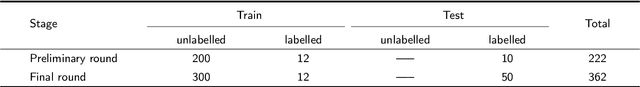
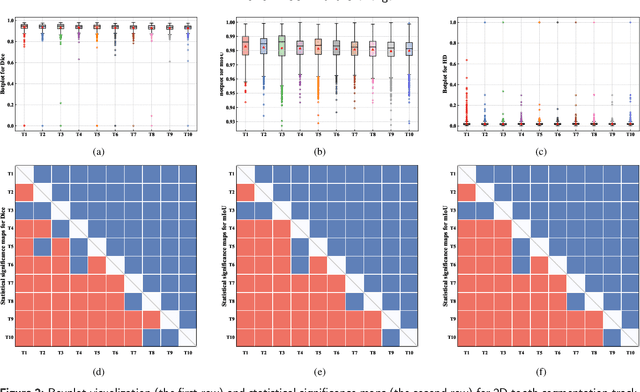
Abstract:Computer-aided design (CAD) tools are increasingly popular in modern dental practice, particularly for treatment planning or comprehensive prognosis evaluation. In particular, the 2D panoramic X-ray image efficiently detects invisible caries, impacted teeth and supernumerary teeth in children, while the 3D dental cone beam computed tomography (CBCT) is widely used in orthodontics and endodontics due to its low radiation dose. However, there is no open-access 2D public dataset for children's teeth and no open 3D dental CBCT dataset, which limits the development of automatic algorithms for segmenting teeth and analyzing diseases. The Semi-supervised Teeth Segmentation (STS) Challenge, a pioneering event in tooth segmentation, was held as a part of the MICCAI 2023 ToothFairy Workshop on the Alibaba Tianchi platform. This challenge aims to investigate effective semi-supervised tooth segmentation algorithms to advance the field of dentistry. In this challenge, we provide two modalities including the 2D panoramic X-ray images and the 3D CBCT tooth volumes. In Task 1, the goal was to segment tooth regions in panoramic X-ray images of both adult and pediatric teeth. Task 2 involved segmenting tooth sections using CBCT volumes. Limited labelled images with mostly unlabelled ones were provided in this challenge prompt using semi-supervised algorithms for training. In the preliminary round, the challenge received registration and result submission by 434 teams, with 64 advancing to the final round. This paper summarizes the diverse methods employed by the top-ranking teams in the STS MICCAI 2023 Challenge.
Hierarchical Point-based Active Learning for Semi-supervised Point Cloud Semantic Segmentation
Aug 22, 2023



Abstract:Impressive performance on point cloud semantic segmentation has been achieved by fully-supervised methods with large amounts of labelled data. As it is labour-intensive to acquire large-scale point cloud data with point-wise labels, many attempts have been made to explore learning 3D point cloud segmentation with limited annotations. Active learning is one of the effective strategies to achieve this purpose but is still under-explored. The most recent methods of this kind measure the uncertainty of each pre-divided region for manual labelling but they suffer from redundant information and require additional efforts for region division. This paper aims at addressing this issue by developing a hierarchical point-based active learning strategy. Specifically, we measure the uncertainty for each point by a hierarchical minimum margin uncertainty module which considers the contextual information at multiple levels. Then, a feature-distance suppression strategy is designed to select important and representative points for manual labelling. Besides, to better exploit the unlabelled data, we build a semi-supervised segmentation framework based on our active strategy. Extensive experiments on the S3DIS and ScanNetV2 datasets demonstrate that the proposed framework achieves 96.5% and 100% performance of fully-supervised baseline with only 0.07% and 0.1% training data, respectively, outperforming the state-of-the-art weakly-supervised and active learning methods. The code will be available at https://github.com/SmiletoE/HPAL.
Joint Dense-Point Representation for Contour-Aware Graph Segmentation
Jun 21, 2023Abstract:We present a novel methodology that combines graph and dense segmentation techniques by jointly learning both point and pixel contour representations, thereby leveraging the benefits of each approach. This addresses deficiencies in typical graph segmentation methods where misaligned objectives restrict the network from learning discriminative vertex and contour features. Our joint learning strategy allows for rich and diverse semantic features to be encoded, while alleviating common contour stability issues in dense-based approaches, where pixel-level objectives can lead to anatomically implausible topologies. In addition, we identify scenarios where correct predictions that fall on the contour boundary are penalised and address this with a novel hybrid contour distance loss. Our approach is validated on several Chest X-ray datasets, demonstrating clear improvements in segmentation stability and accuracy against a variety of dense- and point-based methods. Our source code is freely available at: www.github.com/kitbransby/Joint_Graph_Segmentation
3D Coronary Vessel Reconstruction from Bi-Plane Angiography using Graph Convolutional Networks
Feb 28, 2023
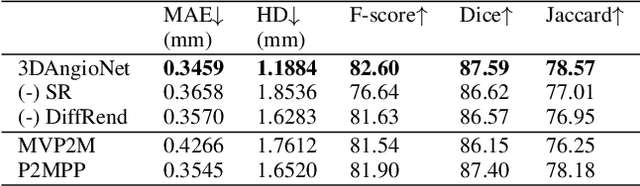


Abstract:X-ray coronary angiography (XCA) is used to assess coronary artery disease and provides valuable information on lesion morphology and severity. However, XCA images are 2D and therefore limit visualisation of the vessel. 3D reconstruction of coronary vessels is possible using multiple views, however lumen border detection in current software is performed manually resulting in limited reproducibility and slow processing time. In this study we propose 3DAngioNet, a novel deep learning (DL) system that enables rapid 3D vessel mesh reconstruction using 2D XCA images from two views. Our approach learns a coarse mesh template using an EfficientB3-UNet segmentation network and projection geometries, and deforms it using a graph convolutional network. 3DAngioNet outperforms similar automated reconstruction methods, offers improved efficiency, and enables modelling of bifurcated vessels. The approach was validated using state-of-the-art software verified by skilled cardiologists.
CTooth+: A Large-scale Dental Cone Beam Computed Tomography Dataset and Benchmark for Tooth Volume Segmentation
Aug 02, 2022
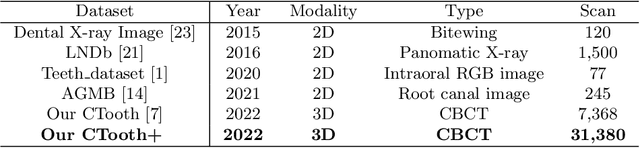
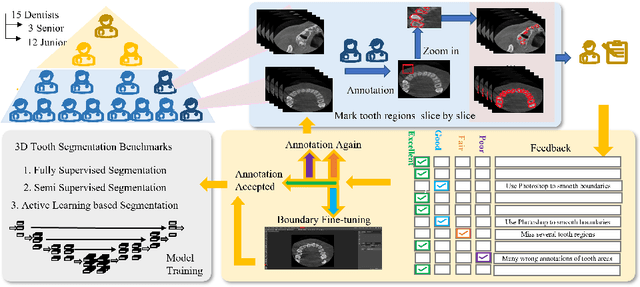
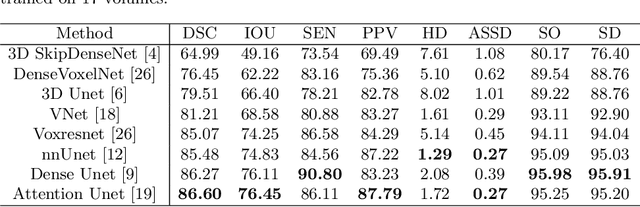
Abstract:Accurate tooth volume segmentation is a prerequisite for computer-aided dental analysis. Deep learning-based tooth segmentation methods have achieved satisfying performances but require a large quantity of tooth data with ground truth. The dental data publicly available is limited meaning the existing methods can not be reproduced, evaluated and applied in clinical practice. In this paper, we establish a 3D dental CBCT dataset CTooth+, with 22 fully annotated volumes and 146 unlabeled volumes. We further evaluate several state-of-the-art tooth volume segmentation strategies based on fully-supervised learning, semi-supervised learning and active learning, and define the performance principles. This work provides a new benchmark for the tooth volume segmentation task, and the experiment can serve as the baseline for future AI-based dental imaging research and clinical application development.
DU-Net based Unsupervised Contrastive Learning for Cancer Segmentation in Histology Images
Jun 17, 2022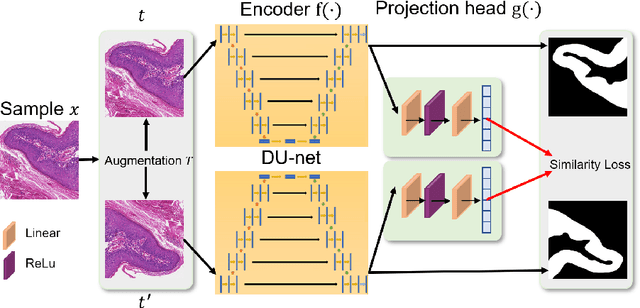
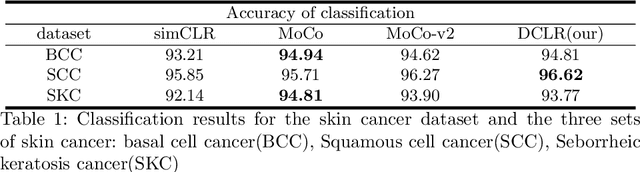
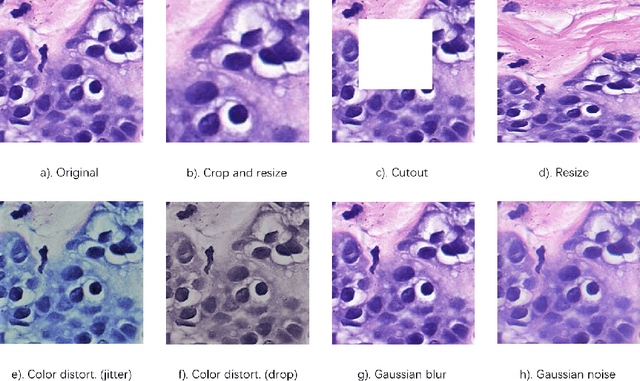
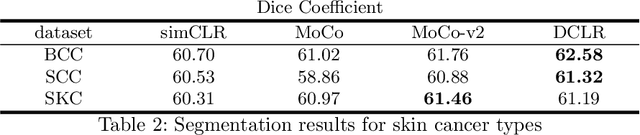
Abstract:In this paper, we introduce an unsupervised cancer segmentation framework for histology images. The framework involves an effective contrastive learning scheme for extracting distinctive visual representations for segmentation. The encoder is a Deep U-Net (DU-Net) structure that contains an extra fully convolution layer compared to the normal U-Net. A contrastive learning scheme is developed to solve the problem of lacking training sets with high-quality annotations on tumour boundaries. A specific set of data augmentation techniques are employed to improve the discriminability of the learned colour features from contrastive learning. Smoothing and noise elimination are conducted using convolutional Conditional Random Fields. The experiments demonstrate competitive performance in segmentation even better than some popular supervised networks.
CTooth: A Fully Annotated 3D Dataset and Benchmark for Tooth Volume Segmentation on Cone Beam Computed Tomography Images
Jun 17, 2022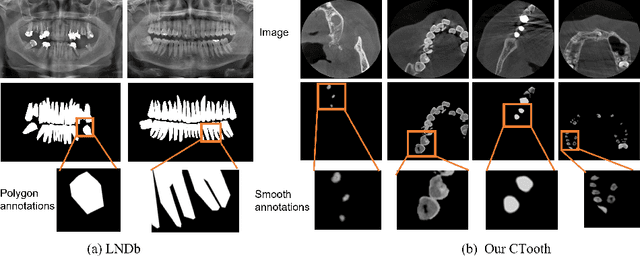
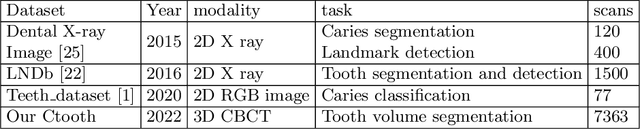
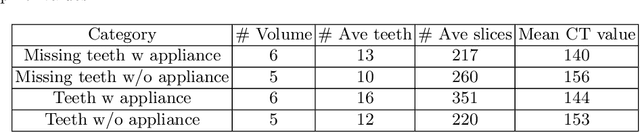
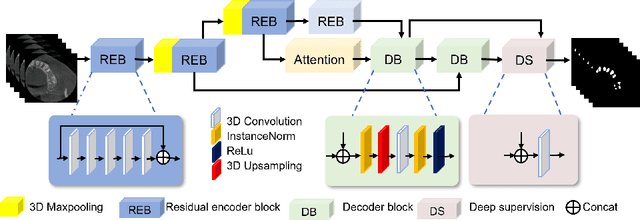
Abstract:3D tooth segmentation is a prerequisite for computer-aided dental diagnosis and treatment. However, segmenting all tooth regions manually is subjective and time-consuming. Recently, deep learning-based segmentation methods produce convincing results and reduce manual annotation efforts, but it requires a large quantity of ground truth for training. To our knowledge, there are few tooth data available for the 3D segmentation study. In this paper, we establish a fully annotated cone beam computed tomography dataset CTooth with tooth gold standard. This dataset contains 22 volumes (7363 slices) with fine tooth labels annotated by experienced radiographic interpreters. To ensure a relative even data sampling distribution, data variance is included in the CTooth including missing teeth and dental restoration. Several state-of-the-art segmentation methods are evaluated on this dataset. Afterwards, we further summarise and apply a series of 3D attention-based Unet variants for segmenting tooth volumes. This work provides a new benchmark for the tooth volume segmentation task. Experimental evidence proves that attention modules of the 3D UNet structure boost responses in tooth areas and inhibit the influence of background and noise. The best performance is achieved by 3D Unet with SKNet attention module, of 88.04 \% Dice and 78.71 \% IOU, respectively. The attention-based Unet framework outperforms other state-of-the-art methods on the CTooth dataset. The codebase and dataset are released.
Complexity Reduction of Learned In-Loop Filtering in Video Coding
Mar 17, 2022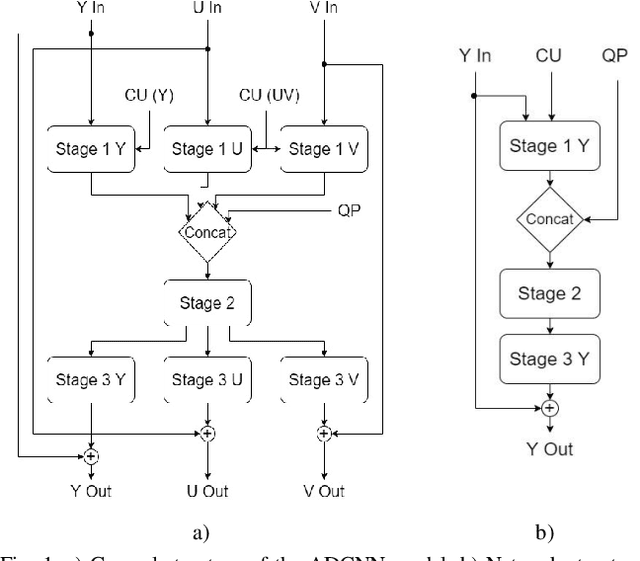
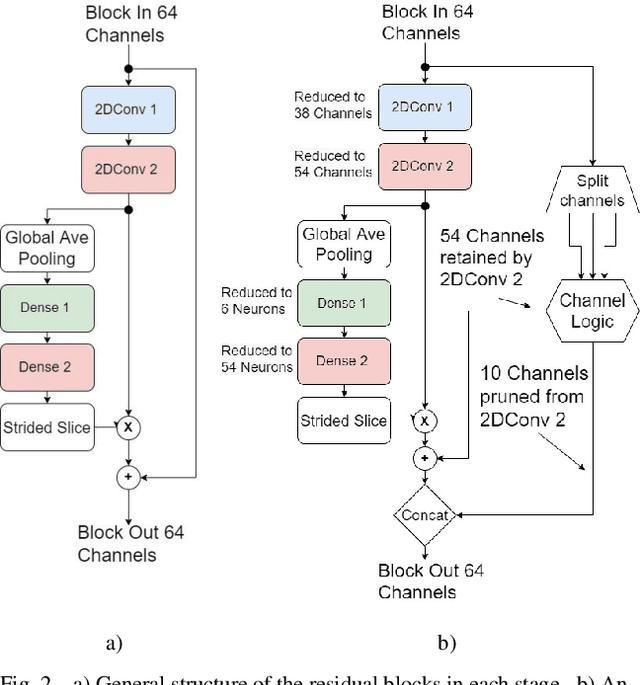
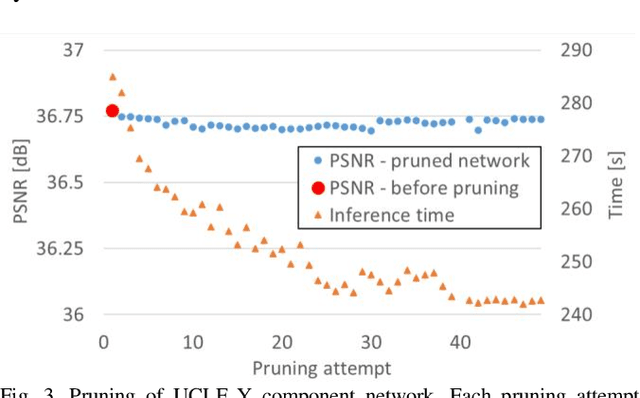
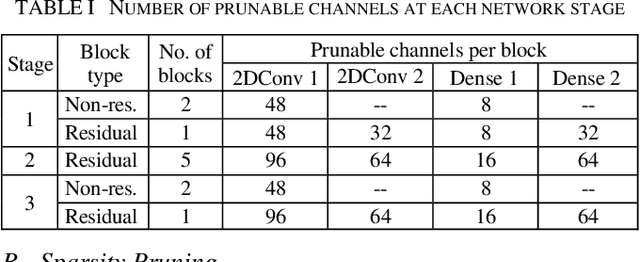
Abstract:In video coding, in-loop filters are applied on reconstructed video frames to enhance their perceptual quality, before storing the frames for output. Conventional in-loop filters are obtained by hand-crafted methods. Recently, learned filters based on convolutional neural networks that utilize attention mechanisms have been shown to improve upon traditional techniques. However, these solutions are typically significantly more computationally expensive, limiting their potential for practical applications. The proposed method uses a novel combination of sparsity and structured pruning for complexity reduction of learned in-loop filters. This is done through a three-step training process of magnitude-guidedweight pruning, insignificant neuron identification and removal, and fine-tuning. Through initial tests we find that network parameters can be significantly reduced with a minimal impact on network performance.
Dispensed Transformer Network for Unsupervised Domain Adaptation
Oct 28, 2021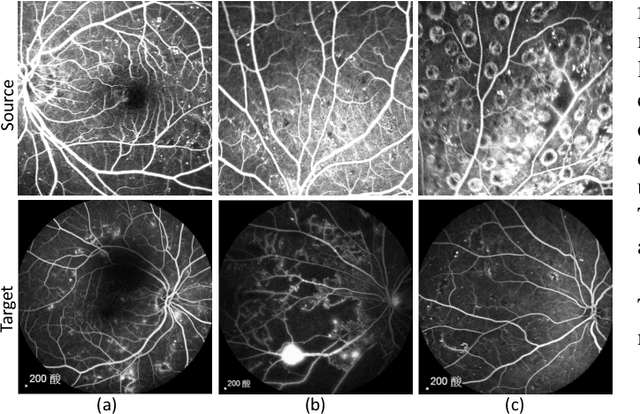
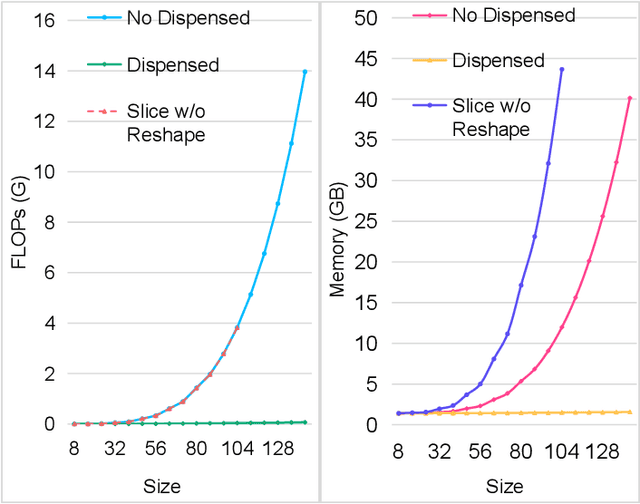
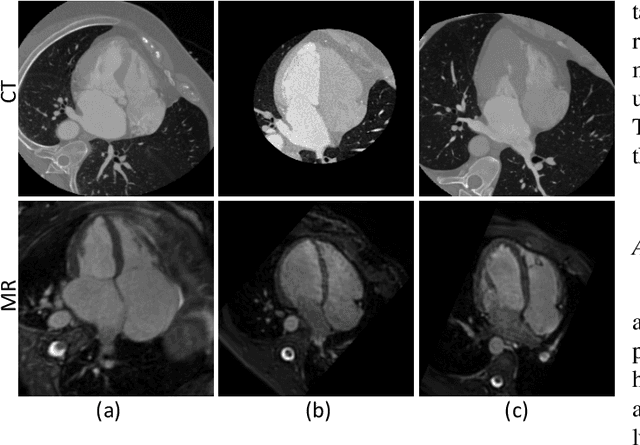
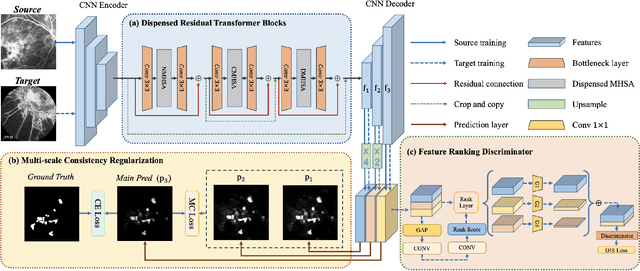
Abstract:Accurate segmentation is a crucial step in medical image analysis and applying supervised machine learning to segment the organs or lesions has been substantiated effective. However, it is costly to perform data annotation that provides ground truth labels for training the supervised algorithms, and the high variance of data that comes from different domains tends to severely degrade system performance over cross-site or cross-modality datasets. To mitigate this problem, a novel unsupervised domain adaptation (UDA) method named dispensed Transformer network (DTNet) is introduced in this paper. Our novel DTNet contains three modules. First, a dispensed residual transformer block is designed, which realizes global attention by dispensed interleaving operation and deals with the excessive computational cost and GPU memory usage of the Transformer. Second, a multi-scale consistency regularization is proposed to alleviate the loss of details in the low-resolution output for better feature alignment. Finally, a feature ranking discriminator is introduced to automatically assign different weights to domain-gap features to lessen the feature distribution distance, reducing the performance shift of two domains. The proposed method is evaluated on large fluorescein angiography (FA) retinal nonperfusion (RNP) cross-site dataset with 676 images and a wide used cross-modality dataset from the MM-WHS challenge. Extensive results demonstrate that our proposed network achieves the best performance in comparison with several state-of-the-art techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge