Guodong Zeng
A Noise-level-aware Framework for PET Image Denoising
Mar 15, 2022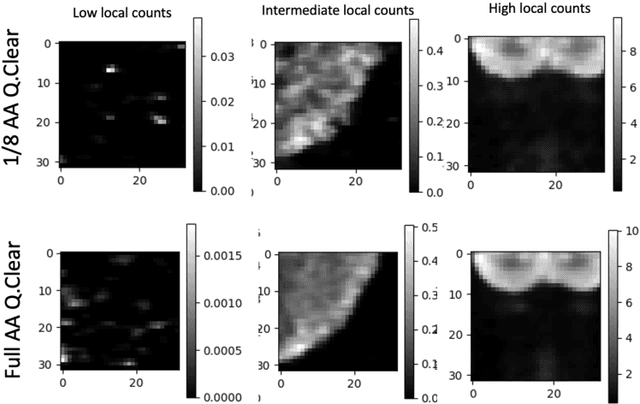
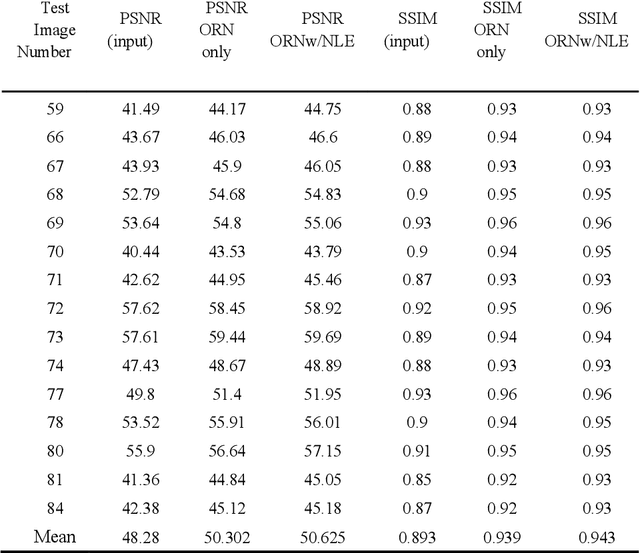
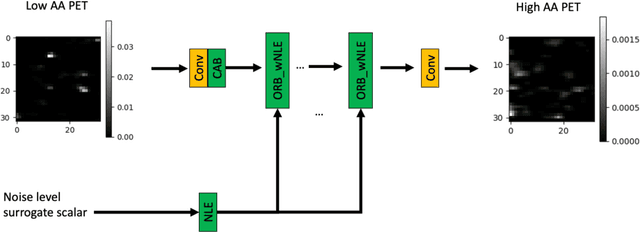
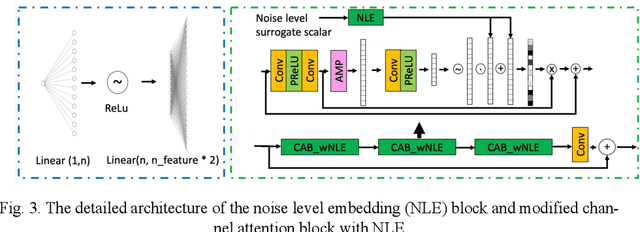
Abstract:In PET, the amount of relative (signal-dependent) noise present in different body regions can be significantly different and is inherently related to the number of counts present in that region. The number of counts in a region depends, in principle and among other factors, on the total administered activity, scanner sensitivity, image acquisition duration, radiopharmaceutical tracer uptake in the region, and patient local body morphometry surrounding the region. In theory, less amount of denoising operations is needed to denoise a high-count (low relative noise) image than images a low-count (high relative noise) image, and vice versa. The current deep-learning-based methods for PET image denoising are predominantly trained on image appearance only and have no special treatment for images of different noise levels. Our hypothesis is that by explicitly providing the local relative noise level of the input image to a deep convolutional neural network (DCNN), the DCNN can outperform itself trained on image appearance only. To this end, we propose a noise-level-aware framework denoising framework that allows embedding of local noise level into a DCNN. The proposed is trained and tested on 30 and 15 patient PET images acquired on a GE Discovery MI PET/CT system. Our experiments showed that the increases in both PSNR and SSIM from our backbone network with relative noise level embedding (NLE) versus the same network without NLE were statistically significant with p<0.001, and the proposed method significantly outperformed a strong baseline method by a large margin.
Dispensed Transformer Network for Unsupervised Domain Adaptation
Oct 28, 2021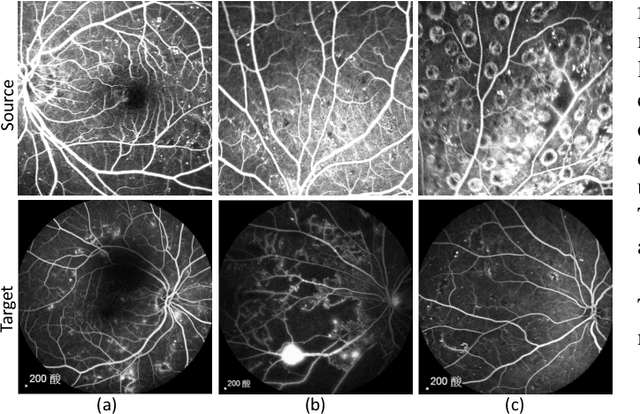
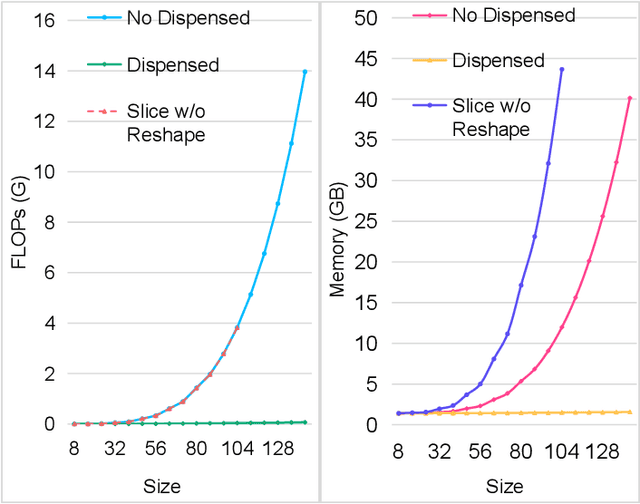
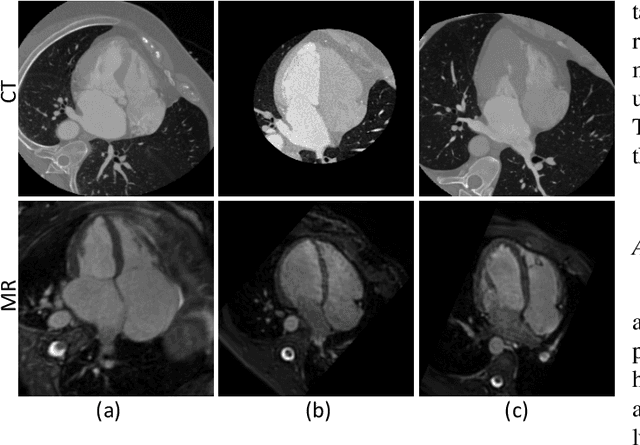
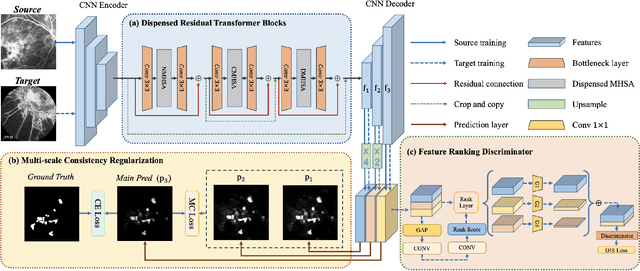
Abstract:Accurate segmentation is a crucial step in medical image analysis and applying supervised machine learning to segment the organs or lesions has been substantiated effective. However, it is costly to perform data annotation that provides ground truth labels for training the supervised algorithms, and the high variance of data that comes from different domains tends to severely degrade system performance over cross-site or cross-modality datasets. To mitigate this problem, a novel unsupervised domain adaptation (UDA) method named dispensed Transformer network (DTNet) is introduced in this paper. Our novel DTNet contains three modules. First, a dispensed residual transformer block is designed, which realizes global attention by dispensed interleaving operation and deals with the excessive computational cost and GPU memory usage of the Transformer. Second, a multi-scale consistency regularization is proposed to alleviate the loss of details in the low-resolution output for better feature alignment. Finally, a feature ranking discriminator is introduced to automatically assign different weights to domain-gap features to lessen the feature distribution distance, reducing the performance shift of two domains. The proposed method is evaluated on large fluorescein angiography (FA) retinal nonperfusion (RNP) cross-site dataset with 676 images and a wide used cross-modality dataset from the MM-WHS challenge. Extensive results demonstrate that our proposed network achieves the best performance in comparison with several state-of-the-art techniques.
GT U-Net: A U-Net Like Group Transformer Network for Tooth Root Segmentation
Sep 30, 2021
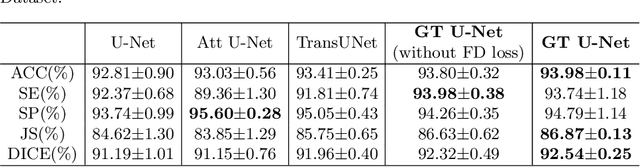

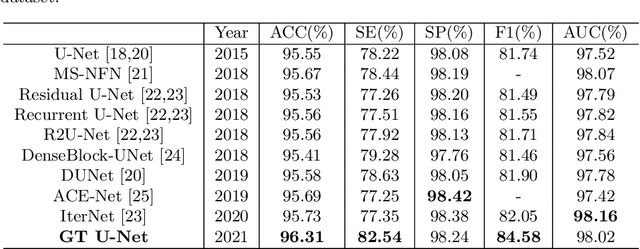
Abstract:To achieve an accurate assessment of root canal therapy, a fundamental step is to perform tooth root segmentation on oral X-ray images, in that the position of tooth root boundary is significant anatomy information in root canal therapy evaluation. However, the fuzzy boundary makes the tooth root segmentation very challenging. In this paper, we propose a novel end-to-end U-Net like Group Transformer Network (GT U-Net) for the tooth root segmentation. The proposed network retains the essential structure of U-Net but each of the encoders and decoders is replaced by a group Transformer, which significantly reduces the computational cost of traditional Transformer architectures by using the grouping structure and the bottleneck structure. In addition, the proposed GT U-Net is composed of a hybrid structure of convolution and Transformer, which makes it independent of pre-training weights. For optimization, we also propose a shape-sensitive Fourier Descriptor (FD) loss function to make use of shape prior knowledge. Experimental results show that our proposed network achieves the state-of-the-art performance on our collected tooth root segmentation dataset and the public retina dataset DRIVE. Code has been released at https://github.com/Kent0n-Li/GT-U-Net.
Anatomy-Guided Parallel Bottleneck Transformer Network for Automated Evaluation of Root Canal Therapy
May 02, 2021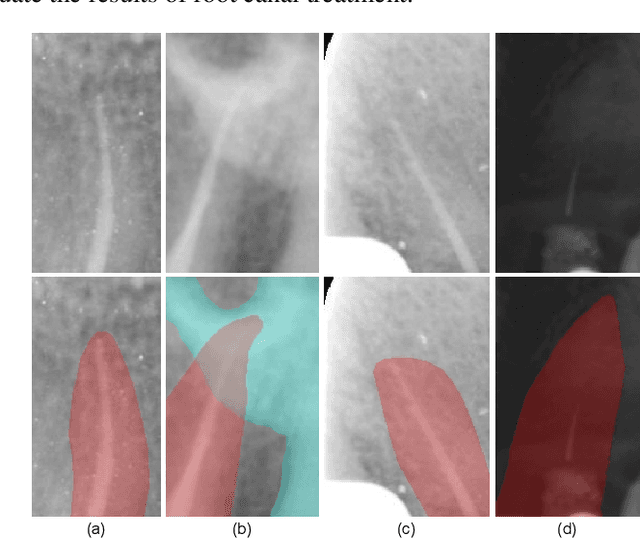
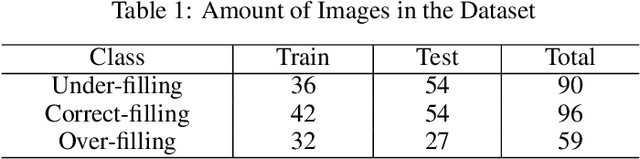
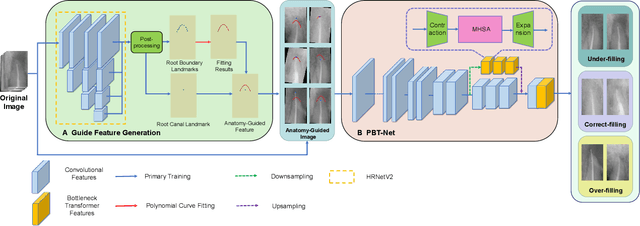
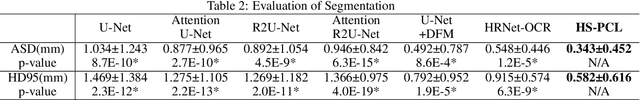
Abstract:Objective: Accurate evaluation of the root canal filling result in X-ray image is a significant step for the root canal therapy, which is based on the relative position between the apical area boundary of tooth root and the top of filled gutta-percha in root canal as well as the shape of the tooth root and so on to classify the result as correct-filling, under-filling or over-filling. Methods: We propose a novel anatomy-guided Transformer diagnosis network. For obtaining accurate anatomy-guided features, a polynomial curve fitting segmentation is proposed to segment the fuzzy boundary. And a Parallel Bottleneck Transformer network (PBT-Net) is introduced as the classification network for the final evaluation. Results, and conclusion: Our numerical experiments show that our anatomy-guided PBT-Net improves the accuracy from 40\% to 85\% relative to the baseline classification network. Comparing with the SOTA segmentation network indicates that the ASD is significantly reduced by 30.3\% through our fitting segmentation. Significance: Polynomial curve fitting segmentation has a great segmentation effect for extremely fuzzy boundaries. The prior knowledge guided classification network is suitable for the evaluation of root canal therapy greatly. And the new proposed Parallel Bottleneck Transformer for realizing self-attention is general in design, facilitating a broad use in most backbone networks.
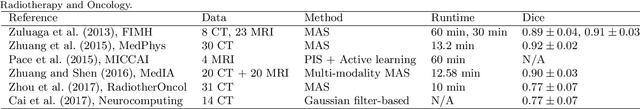
The MICCAI Hackathon on reproducibility, diversity, and selection of papers at the MICCAI conference
Mar 04, 2021
Abstract:The MICCAI conference has encountered tremendous growth over the last years in terms of the size of the community, as well as the number of contributions and their technical success. With this growth, however, come new challenges for the community. Methods are more difficult to reproduce and the ever-increasing number of paper submissions to the MICCAI conference poses new questions regarding the selection process and the diversity of topics. To exchange, discuss, and find novel and creative solutions to these challenges, a new format of a hackathon was initiated as a satellite event at the MICCAI 2020 conference: The MICCAI Hackathon. The first edition of the MICCAI Hackathon covered the topics reproducibility, diversity, and selection of MICCAI papers. In the manner of a small think-tank, participants collaborated to find solutions to these challenges. In this report, we summarize the insights from the MICCAI Hackathon into immediate and long-term measures to address these challenges. The proposed measures can be seen as starting points and guidelines for discussions and actions to possibly improve the MICCAI conference with regards to reproducibility, diversity, and selection of papers.
ICMSC: Intra- and Cross-modality Semantic Consistency for Unsupervised Domain Adaptation on Hip Joint Bone Segmentation
Dec 23, 2020


Abstract:Unsupervised domain adaptation (UDA) for cross-modality medical image segmentation has shown great progress by domain-invariant feature learning or image appearance translation. Adapted feature learning usually cannot detect domain shifts at the pixel level and is not able to achieve good results in dense semantic segmentation tasks. Image appearance translation, e.g. CycleGAN, translates images into different styles with good appearance, despite its population, its semantic consistency is hardly to maintain and results in poor cross-modality segmentation. In this paper, we propose intra- and cross-modality semantic consistency (ICMSC) for UDA and our key insight is that the segmentation of synthesised images in different styles should be consistent. Specifically, our model consists of an image translation module and a domain-specific segmentation module. The image translation module is a standard CycleGAN, while the segmentation module contains two domain-specific segmentation networks. The intra-modality semantic consistency (IMSC) forces the reconstructed image after a cycle to be segmented in the same way as the original input image, while the cross-modality semantic consistency (CMSC) encourages the synthesized images after translation to be segmented exactly the same as before translation. Comprehensive experimental results on cross-modality hip joint bone segmentation show the effectiveness of our proposed method, which achieves an average DICE of 81.61% on the acetabulum and 88.16% on the proximal femur, outperforming other state-of-the-art methods. It is worth to note that without UDA, a model trained on CT for hip joint bone segmentation is non-transferable to MRI and has almost zero-DICE segmentation.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019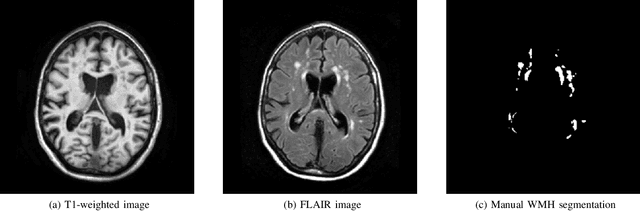
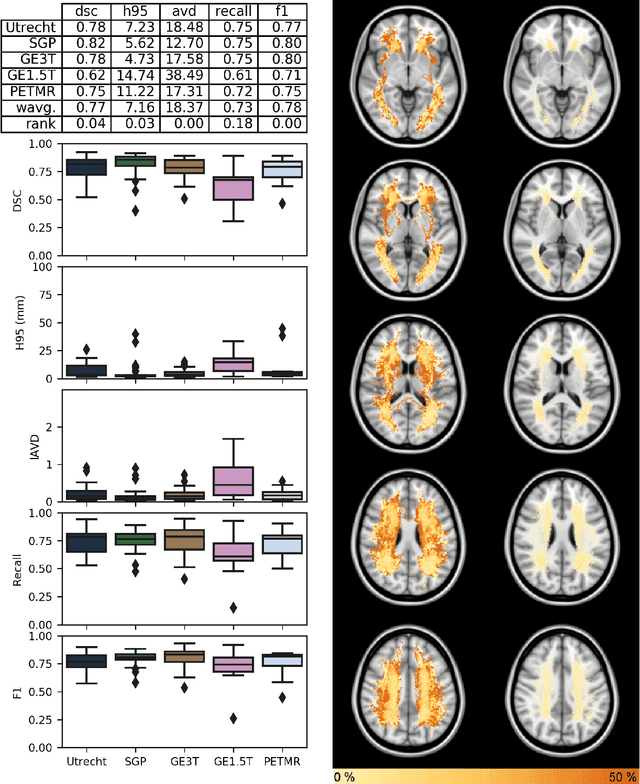
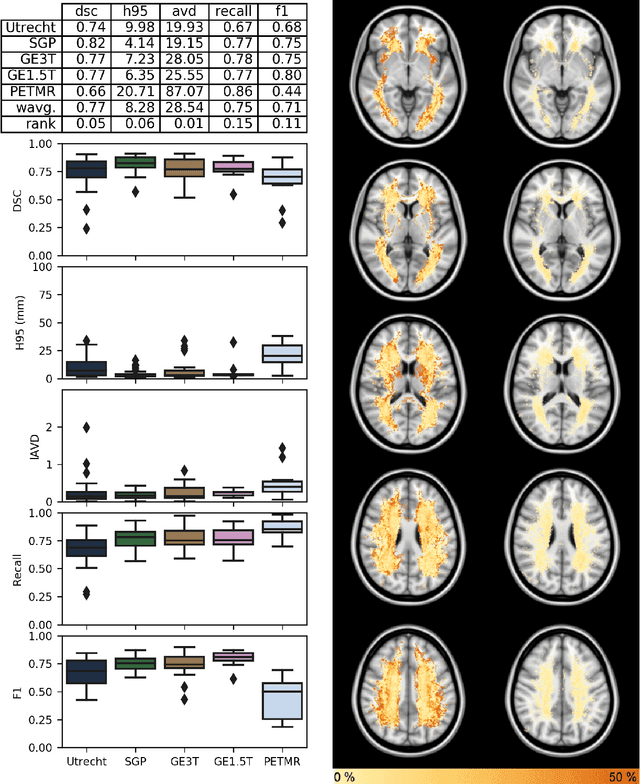
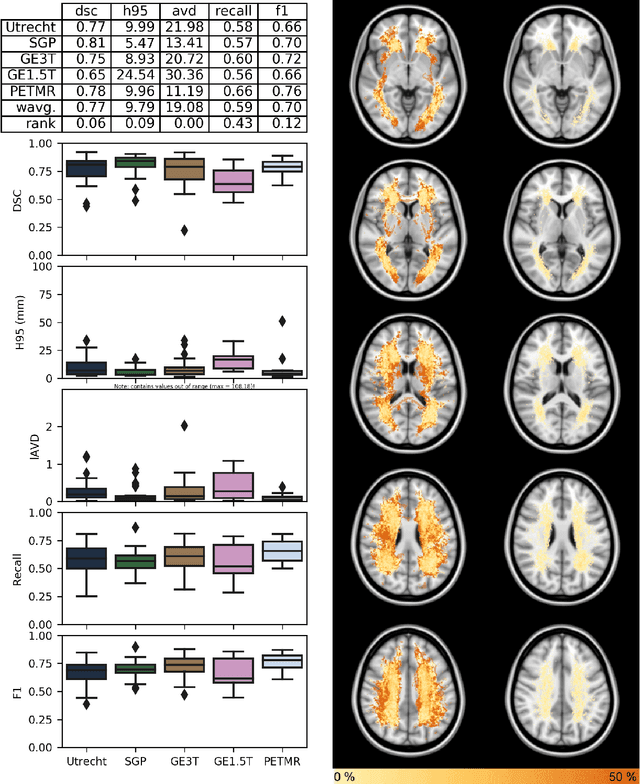
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019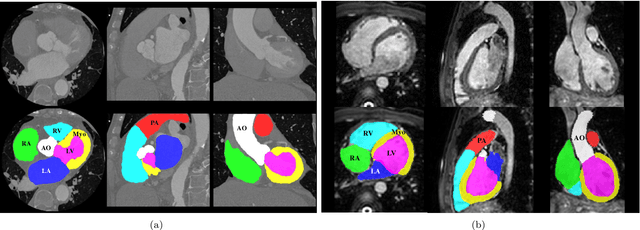
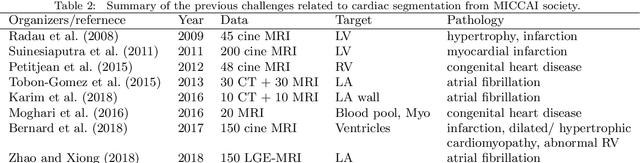
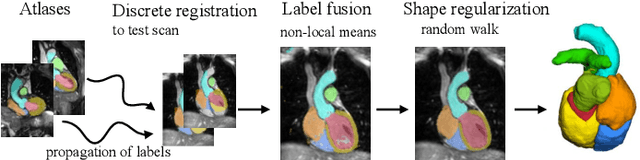
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
Holistic Decomposition Convolution for Effective Semantic Segmentation of 3D MR Images
Dec 24, 2018



Abstract:Convolutional Neural Networks (CNNs) have achieved state-of-the-art performance in many different 2D medical image analysis tasks. In clinical practice, however, a large part of the medical imaging data available is in 3D. This has motivated the development of 3D CNNs for volumetric image segmentation in order to benefit from more spatial context. Due to GPU memory restrictions caused by moving to fully 3D, state-of-the-art methods depend on subvolume/patch processing and the size of the input patch is usually small, limiting the incorporation of larger context information for a better performance. In this paper, we propose a novel Holistic Decomposition Convolution (HDC), for an effective and efficient semantic segmentation of volumetric images. HDC consists of a periodic down-shuffling operation followed by a conventional 3D convolution. HDC has the advantage of significantly reducing the size of the data for sub-sequential processing while using all the information available in the input irrespective of the down-shuffling factors. Results obtained from comprehensive experiments conducted on hip T1 MR images and intervertebral disc T2 MR images demonstrate the efficacy of the present approach.
Fully Automatic Segmentation of Lumbar Vertebrae from CT Images using Cascaded 3D Fully Convolutional Networks
Dec 05, 2017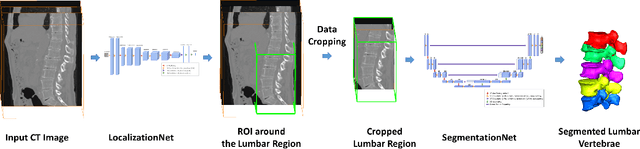
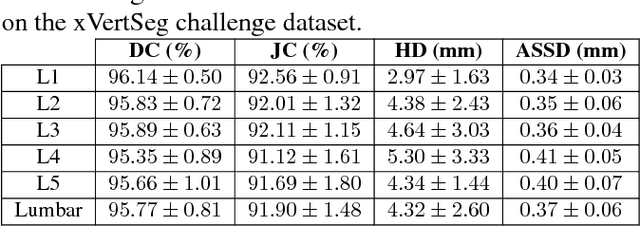
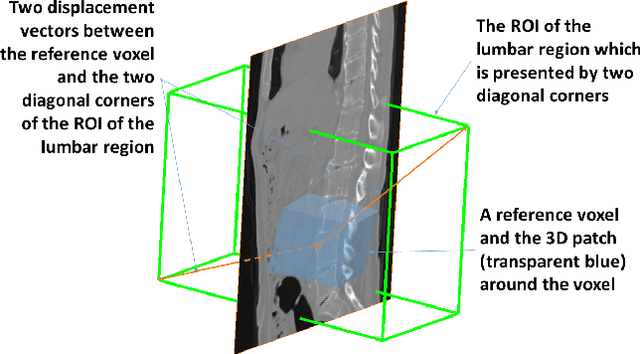
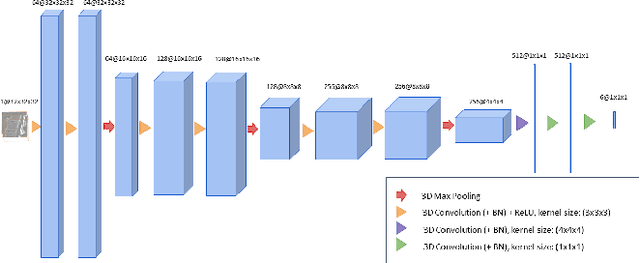
Abstract:We present a method to address the challenging problem of segmentation of lumbar vertebrae from CT images acquired with varying fields of view. Our method is based on cascaded 3D Fully Convolutional Networks (FCNs) consisting of a localization FCN and a segmentation FCN. More specifically, in the first step we train a regression 3D FCN (we call it "LocalizationNet") to find the bounding box of the lumbar region. After that, a 3D U-net like FCN (we call it "SegmentationNet") is then developed, which after training, can perform a pixel-wise multi-class segmentation to map a cropped lumber region volumetric data to its volume-wise labels. Evaluated on publicly available datasets, our method achieved an average Dice coefficient of 95.77 $\pm$ 0.81% and an average symmetric surface distance of 0.37 $\pm$ 0.06 mm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge