Qun Jin
Statistical-Neural Interaction Networks for Interpretable Mixed-Type Data Imputation
Jan 18, 2026Abstract:Real-world tabular databases routinely combine continuous measurements and categorical records, yet missing entries are pervasive and can distort downstream analysis. We propose Statistical-Neural Interaction (SNI), an interpretable mixed-type imputation framework that couples correlation-derived statistical priors with neural feature attention through a Controllable-Prior Feature Attention (CPFA) module. CPFA learns head-wise prior-strength coefficients $\{λ_h\}$ that softly regularize attention toward the prior while allowing data-driven deviations when nonlinear patterns appear to be present in the data. Beyond imputation, SNI aggregates attention maps into a directed feature-dependency matrix that summarizes which variables the imputer relied on, without requiring post-hoc explainers. We evaluate SNI against six baselines (Mean/Mode, MICE, KNN, MissForest, GAIN, MIWAE) on six datasets spanning ICU monitoring, population surveys, socio-economic statistics, and engineering applications. Under MCAR/strict-MAR at 30\% missingness, SNI is generally competitive on continuous metrics but is often outperformed by accuracy-first baselines (MissForest, MIWAE) on categorical variables; in return, it provides intrinsic dependency diagnostics and explicit statistical-neural trade-off parameters. We additionally report MNAR stress tests (with a mask-aware variant) and discuss computational cost, limitations -- particularly for severely imbalanced categorical targets -- and deployment scenarios where interpretability may justify the trade-off.
Phenome-wide causal proteomics enhance systemic lupus erythematosus flare prediction: A study in Asian populations
Nov 18, 2024Abstract:Objective: Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterized by unpredictable flares. This study aimed to develop a novel proteomics-based risk prediction model specifically for Asian SLE populations to enhance personalized disease management and early intervention. Methods: A longitudinal cohort study was conducted over 48 weeks, including 139 SLE patients monitored every 12 weeks. Patients were classified into flare (n = 53) and non-flare (n = 86) groups. Baseline plasma samples underwent data-independent acquisition (DIA) proteomics analysis, and phenome-wide Mendelian randomization (PheWAS) was performed to evaluate causal relationships between proteins and clinical predictors. Logistic regression (LR) and random forest (RF) models were used to integrate proteomic and clinical data for flare risk prediction. Results: Five proteins (SAA1, B4GALT5, GIT2, NAA15, and RPIA) were significantly associated with SLE Disease Activity Index-2K (SLEDAI-2K) scores and 1-year flare risk, implicating key pathways such as B-cell receptor signaling and platelet degranulation. SAA1 demonstrated causal effects on flare-related clinical markers, including hemoglobin and red blood cell counts. A combined model integrating clinical and proteomic data achieved the highest predictive accuracy (AUC = 0.769), surpassing individual models. SAA1 was highlighted as a priority biomarker for rapid flare discrimination. Conclusion: The integration of proteomic and clinical data significantly improves flare prediction in Asian SLE patients. The identification of key proteins and their causal relationships with flare-related clinical markers provides valuable insights for proactive SLE management and personalized therapeutic approaches.
Evolutionary Causal Discovery with Relative Impact Stratification for Interpretable Data Analysis
Apr 25, 2024Abstract:This study proposes Evolutionary Causal Discovery (ECD) for causal discovery that tailors response variables, predictor variables, and corresponding operators to research datasets. Utilizing genetic programming for variable relationship parsing, the method proceeds with the Relative Impact Stratification (RIS) algorithm to assess the relative impact of predictor variables on the response variable, facilitating expression simplification and enhancing the interpretability of variable relationships. ECD proposes an expression tree to visualize the RIS results, offering a differentiated depiction of unknown causal relationships compared to conventional causal discovery. The ECD method represents an evolution and augmentation of existing causal discovery methods, providing an interpretable approach for analyzing variable relationships in complex systems, particularly in healthcare settings with Electronic Health Record (EHR) data. Experiments on both synthetic and real-world EHR datasets demonstrate the efficacy of ECD in uncovering patterns and mechanisms among variables, maintaining high accuracy and stability across different noise levels. On the real-world EHR dataset, ECD reveals the intricate relationships between the response variable and other predictive variables, aligning with the results of structural equation modeling and shapley additive explanations analyses.
FOSA: Full Information Maximum Likelihood (FIML) Optimized Self-Attention Imputation for Missing Data
Aug 23, 2023Abstract:In data imputation, effectively addressing missing values is pivotal, especially in intricate datasets. This paper delves into the FIML Optimized Self-attention (FOSA) framework, an innovative approach that amalgamates the strengths of Full Information Maximum Likelihood (FIML) estimation with the capabilities of self-attention neural networks. Our methodology commences with an initial estimation of missing values via FIML, subsequently refining these estimates by leveraging the self-attention mechanism. Our comprehensive experiments on both simulated and real-world datasets underscore FOSA's pronounced advantages over traditional FIML techniques, encapsulating facets of accuracy, computational efficiency, and adaptability to diverse data structures. Intriguingly, even in scenarios where the Structural Equation Model (SEM) might be mis-specified, leading to suboptimal FIML estimates, the robust architecture of FOSA's self-attention component adeptly rectifies and optimizes the imputation outcomes. Our empirical tests reveal that FOSA consistently delivers commendable predictions, even in the face of up to 40% random missingness, highlighting its robustness and potential for wide-scale applications in data imputation.
CDNet: Contrastive Disentangled Network for Fine-Grained Image Categorization of Ocular B-Scan Ultrasound
Jun 17, 2022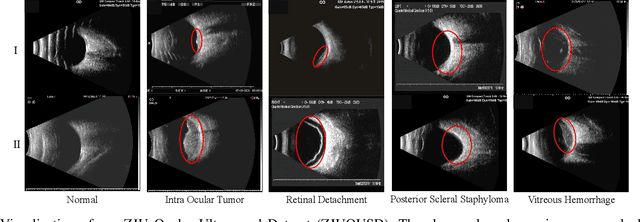
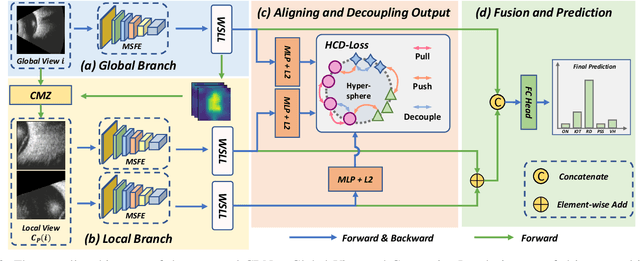
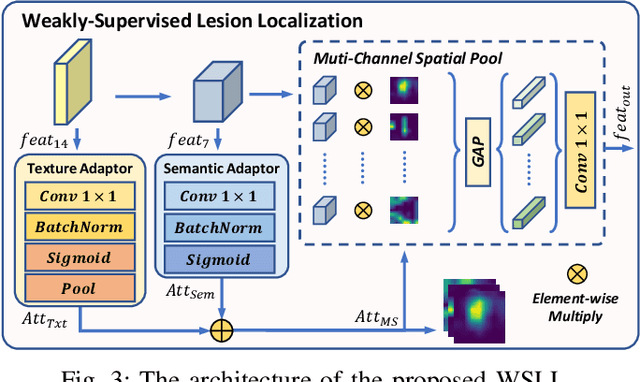
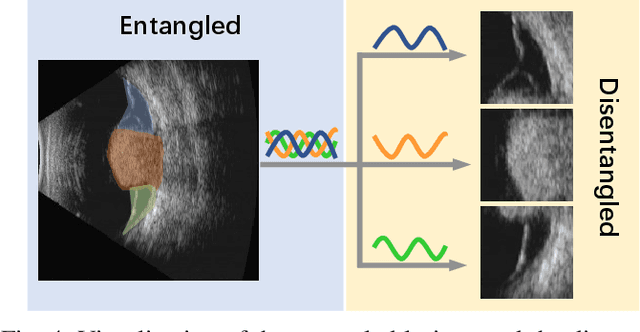
Abstract:Precise and rapid categorization of images in the B-scan ultrasound modality is vital for diagnosing ocular diseases. Nevertheless, distinguishing various diseases in ultrasound still challenges experienced ophthalmologists. Thus a novel contrastive disentangled network (CDNet) is developed in this work, aiming to tackle the fine-grained image categorization (FGIC) challenges of ocular abnormalities in ultrasound images, including intraocular tumor (IOT), retinal detachment (RD), posterior scleral staphyloma (PSS), and vitreous hemorrhage (VH). Three essential components of CDNet are the weakly-supervised lesion localization module (WSLL), contrastive multi-zoom (CMZ) strategy, and hyperspherical contrastive disentangled loss (HCD-Loss), respectively. These components facilitate feature disentanglement for fine-grained recognition in both the input and output aspects. The proposed CDNet is validated on our ZJU Ocular Ultrasound Dataset (ZJUOUSD), consisting of 5213 samples. Furthermore, the generalization ability of CDNet is validated on two public and widely-used chest X-ray FGIC benchmarks. Quantitative and qualitative results demonstrate the efficacy of our proposed CDNet, which achieves state-of-the-art performance in the FGIC task. Code is available at: https://github.com/ZeroOneGame/CDNet-for-OUS-FGIC .
Dispensed Transformer Network for Unsupervised Domain Adaptation
Oct 28, 2021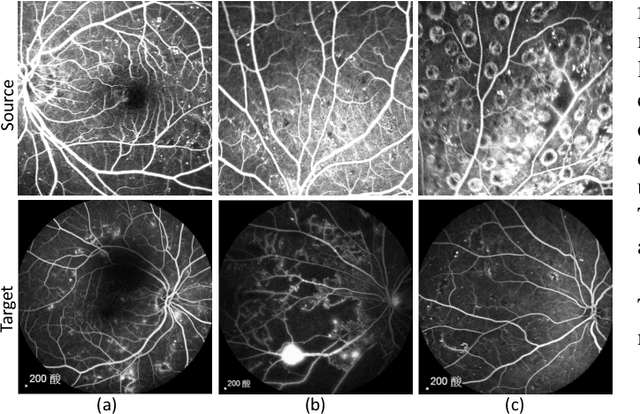
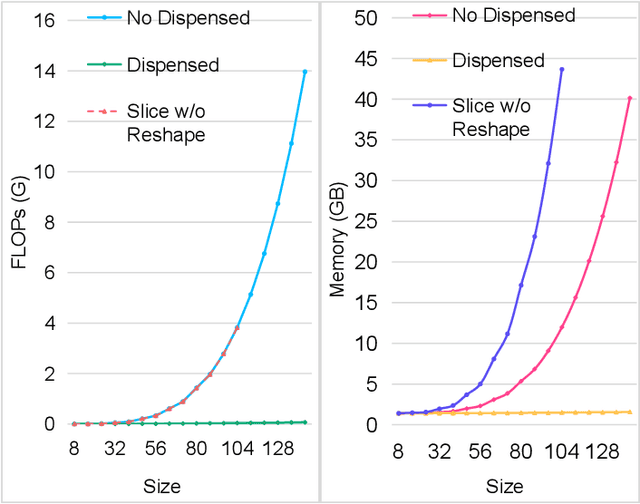
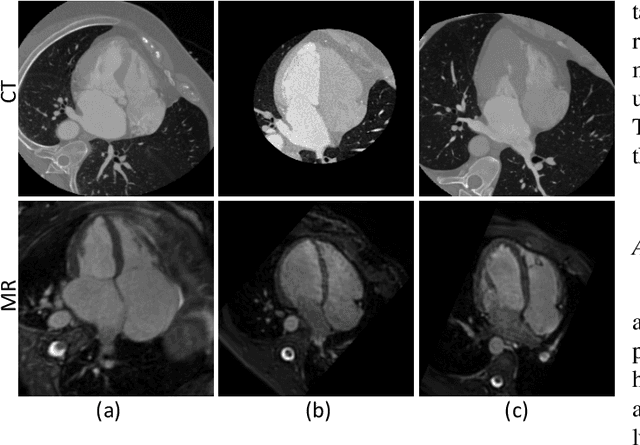
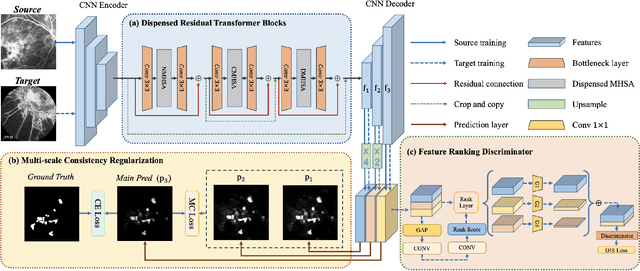
Abstract:Accurate segmentation is a crucial step in medical image analysis and applying supervised machine learning to segment the organs or lesions has been substantiated effective. However, it is costly to perform data annotation that provides ground truth labels for training the supervised algorithms, and the high variance of data that comes from different domains tends to severely degrade system performance over cross-site or cross-modality datasets. To mitigate this problem, a novel unsupervised domain adaptation (UDA) method named dispensed Transformer network (DTNet) is introduced in this paper. Our novel DTNet contains three modules. First, a dispensed residual transformer block is designed, which realizes global attention by dispensed interleaving operation and deals with the excessive computational cost and GPU memory usage of the Transformer. Second, a multi-scale consistency regularization is proposed to alleviate the loss of details in the low-resolution output for better feature alignment. Finally, a feature ranking discriminator is introduced to automatically assign different weights to domain-gap features to lessen the feature distribution distance, reducing the performance shift of two domains. The proposed method is evaluated on large fluorescein angiography (FA) retinal nonperfusion (RNP) cross-site dataset with 676 images and a wide used cross-modality dataset from the MM-WHS challenge. Extensive results demonstrate that our proposed network achieves the best performance in comparison with several state-of-the-art techniques.
GT U-Net: A U-Net Like Group Transformer Network for Tooth Root Segmentation
Sep 30, 2021
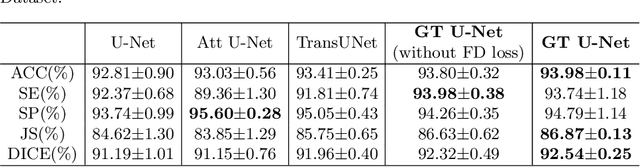

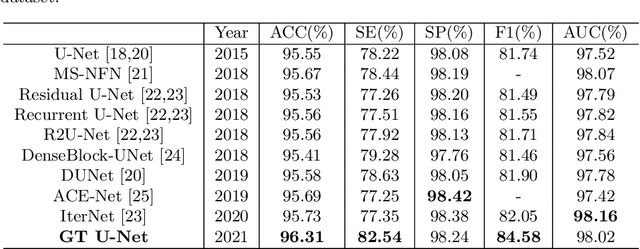
Abstract:To achieve an accurate assessment of root canal therapy, a fundamental step is to perform tooth root segmentation on oral X-ray images, in that the position of tooth root boundary is significant anatomy information in root canal therapy evaluation. However, the fuzzy boundary makes the tooth root segmentation very challenging. In this paper, we propose a novel end-to-end U-Net like Group Transformer Network (GT U-Net) for the tooth root segmentation. The proposed network retains the essential structure of U-Net but each of the encoders and decoders is replaced by a group Transformer, which significantly reduces the computational cost of traditional Transformer architectures by using the grouping structure and the bottleneck structure. In addition, the proposed GT U-Net is composed of a hybrid structure of convolution and Transformer, which makes it independent of pre-training weights. For optimization, we also propose a shape-sensitive Fourier Descriptor (FD) loss function to make use of shape prior knowledge. Experimental results show that our proposed network achieves the state-of-the-art performance on our collected tooth root segmentation dataset and the public retina dataset DRIVE. Code has been released at https://github.com/Kent0n-Li/GT-U-Net.
Anatomy-Guided Parallel Bottleneck Transformer Network for Automated Evaluation of Root Canal Therapy
May 02, 2021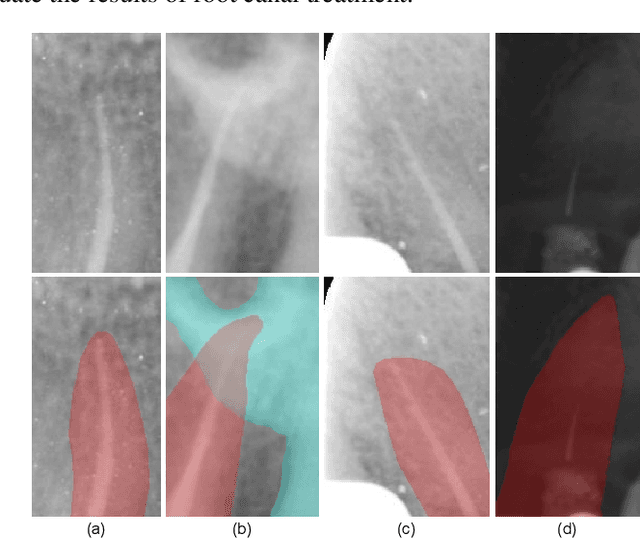
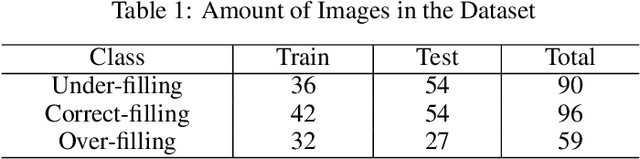
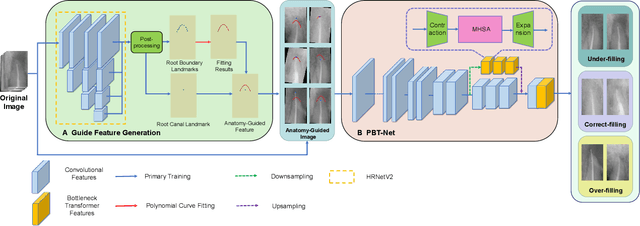
Abstract:Objective: Accurate evaluation of the root canal filling result in X-ray image is a significant step for the root canal therapy, which is based on the relative position between the apical area boundary of tooth root and the top of filled gutta-percha in root canal as well as the shape of the tooth root and so on to classify the result as correct-filling, under-filling or over-filling. Methods: We propose a novel anatomy-guided Transformer diagnosis network. For obtaining accurate anatomy-guided features, a polynomial curve fitting segmentation is proposed to segment the fuzzy boundary. And a Parallel Bottleneck Transformer network (PBT-Net) is introduced as the classification network for the final evaluation. Results, and conclusion: Our numerical experiments show that our anatomy-guided PBT-Net improves the accuracy from 40\% to 85\% relative to the baseline classification network. Comparing with the SOTA segmentation network indicates that the ASD is significantly reduced by 30.3\% through our fitting segmentation. Significance: Polynomial curve fitting segmentation has a great segmentation effect for extremely fuzzy boundaries. The prior knowledge guided classification network is suitable for the evaluation of root canal therapy greatly. And the new proposed Parallel Bottleneck Transformer for realizing self-attention is general in design, facilitating a broad use in most backbone networks.
High-Resolution Segmentation of Tooth Root Fuzzy Edge Based on Polynomial Curve Fitting with Landmark Detection
Mar 07, 2021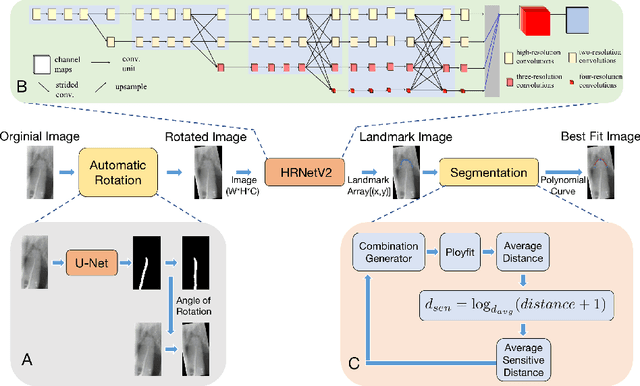

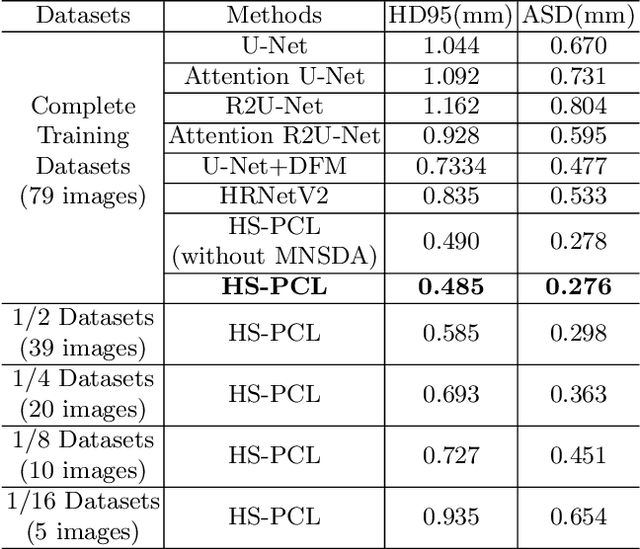
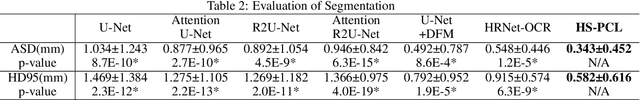
Abstract:As the most economical and routine auxiliary examination in the diagnosis of root canal treatment, oral X-ray has been widely used by stomatologists. It is still challenging to segment the tooth root with a blurry boundary for the traditional image segmentation method. To this end, we propose a model for high-resolution segmentation based on polynomial curve fitting with landmark detection (HS-PCL). It is based on detecting multiple landmarks evenly distributed on the edge of the tooth root to fit a smooth polynomial curve as the segmentation of the tooth root, thereby solving the problem of fuzzy edge. In our model, a maximum number of the shortest distances algorithm (MNSDA) is proposed to automatically reduce the negative influence of the wrong landmarks which are detected incorrectly and deviate from the tooth root on the fitting result. Our numerical experiments demonstrate that the proposed approach not only reduces Hausdorff95 (HD95) by 33.9% and Average Surface Distance (ASD) by 42.1% compared with the state-of-the-art method, but it also achieves excellent results on the minute quantity of datasets, which greatly improves the feasibility of automatic root canal therapy evaluation by medical image computing.
Multiscale Attention Guided Network for COVID-19 Detection Using Chest X-ray Images
Nov 11, 2020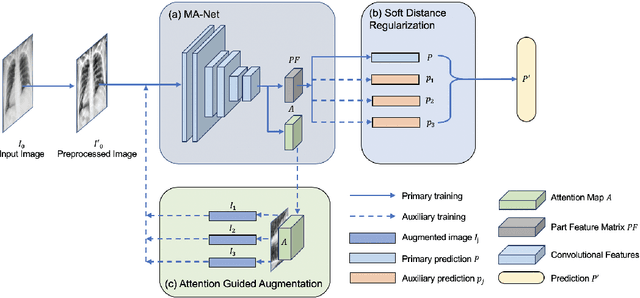
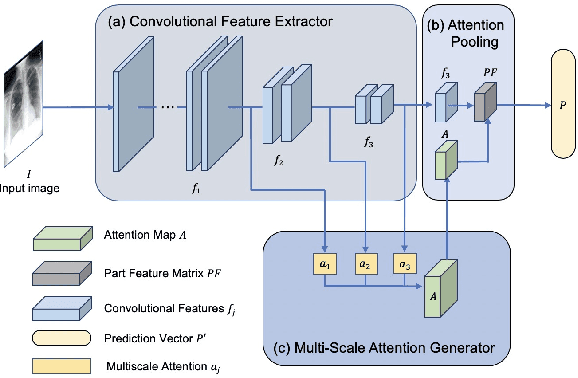
Abstract:Coronavirus disease 2019 (COVID-19) is one of the most destructive pandemic after millennium, forcing the world to tackle a health crisis. Automated classification of lung infections from chest X-ray (CXR) images strengthened traditional healthcare strategy to handle COVID-19. However, classifying COVID-19 from pneumonia cases using CXR image is challenging because of shared spatial characteristics, high feature variation in infections and contrast diversity between cases. Moreover, massive data collection is impractical for a newly emerged disease, which limited the performance of common deep learning models. To address this challenging topic, Multiscale Attention Guided deep network with Soft Distance regularization (MAG-SD) is proposed to automatically classify COVID-19 from pneumonia CXR images. In MAG-SD, MA-Net is used to produce prediction vector and attention map from multiscale feature maps. To relieve the shortage of training data, attention guided augmentations along with a soft distance regularization are posed, which requires a few labeled data to generate meaningful augmentations and reduce noise. Our multiscale attention model achieves better classification performance on our pneumonia CXR image dataset. Plentiful experiments are proposed for MAG-SD which demonstrates that it has its unique advantage in pneumonia classification over cuttingedge models. The code is available at https://github.com/ JasonLeeGHub/MAG-SD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge