Yaqi Wang
VLM-CAD: VLM-Optimized Collaborative Agent Design Workflow for Analog Circuit Sizing
Jan 12, 2026Abstract:Analog mixed-signal circuit sizing involves complex trade-offs within high-dimensional design spaces. Existing automatic analog circuit sizing approaches often underutilize circuit schematics and lack the explainability required for industry adoption. To tackle these challenges, we propose a Vision Language Model-optimized collaborative agent design workflow (VLM-CAD), which analyzes circuits, optimizes DC operating points, performs inference-based sizing and executes external sizing optimization. We integrate Image2Net to annotate circuit schematics and generate a structured JSON description for precise interpretation by Vision Language Models. Furthermore, we propose an Explainable Trust Region Bayesian Optimization method (ExTuRBO) that employs collaborative warm-starting from agent-generated seeds and offers dual-granularity sensitivity analysis for external sizing optimization, supporting a comprehensive final design report. Experiment results on amplifier sizing tasks using 180nm, 90nm, and 45nm Predictive Technology Models demonstrate that VLM-CAD effectively balances power and performance, achieving a 100% success rate in optimizing an amplifier with a complementary input and a class-AB output stage, while maintaining total runtime under 43 minutes across all experiments.
Physically-Grounded Manifold Projection Model for Generalizable Metal Artifact Reduction in Dental CBCT
Jan 01, 2026Abstract:Metal artifacts in Dental CBCT severely obscure anatomical structures, hindering diagnosis. Current deep learning for Metal Artifact Reduction (MAR) faces limitations: supervised methods suffer from spectral blurring due to "regression-to-the-mean", while unsupervised ones risk structural hallucinations. Denoising Diffusion Models (DDPMs) offer realism but rely on slow, stochastic iterative sampling, unsuitable for clinical use. To resolve this, we propose the Physically-Grounded Manifold Projection (PGMP) framework. First, our Anatomically-Adaptive Physics Simulation (AAPS) pipeline synthesizes high-fidelity training pairs via Monte Carlo spectral modeling and patient-specific digital twins, bridging the synthetic-to-real gap. Second, our DMP-Former adapts the Direct x-Prediction paradigm, reformulating restoration as a deterministic manifold projection to recover clean anatomy in a single forward pass, eliminating stochastic sampling. Finally, a Semantic-Structural Alignment (SSA) module anchors the solution using priors from medical foundation models (MedDINOv3), ensuring clinical plausibility. Experiments on synthetic and multi-center clinical datasets show PGMP outperforms state-of-the-art methods on unseen anatomy, setting new benchmarks in efficiency and diagnostic reliability. Code and data: https://github.com/ricoleehduu/PGMP.
Physically-Grounded Manifold Projection with Foundation Priors for Metal Artifact Reduction in Dental CBCT
Dec 30, 2025Abstract:Metal artifacts in Dental CBCT severely obscure anatomical structures, hindering diagnosis. Current deep learning for Metal Artifact Reduction (MAR) faces limitations: supervised methods suffer from spectral blurring due to "regression-to-the-mean", while unsupervised ones risk structural hallucinations. Denoising Diffusion Models (DDPMs) offer realism but rely on slow, stochastic iterative sampling, unsuitable for clinical use. To resolve this, we propose the Physically-Grounded Manifold Projection (PGMP) framework. First, our Anatomically-Adaptive Physics Simulation (AAPS) pipeline synthesizes high-fidelity training pairs via Monte Carlo spectral modeling and patient-specific digital twins, bridging the synthetic-to-real gap. Second, our DMP-Former adapts the Direct x-Prediction paradigm, reformulating restoration as a deterministic manifold projection to recover clean anatomy in a single forward pass, eliminating stochastic sampling. Finally, a Semantic-Structural Alignment (SSA) module anchors the solution using priors from medical foundation models (MedDINOv3), ensuring clinical plausibility. Experiments on synthetic and multi-center clinical datasets show PGMP outperforms state-of-the-art methods on unseen anatomy, setting new benchmarks in efficiency and diagnostic reliability. Code and data: https://github.com/ricoleehduu/PGMP
SASG-DA: Sparse-Aware Semantic-Guided Diffusion Augmentation For Myoelectric Gesture Recognition
Nov 12, 2025Abstract:Surface electromyography (sEMG)-based gesture recognition plays a critical role in human-machine interaction (HMI), particularly for rehabilitation and prosthetic control. However, sEMG-based systems often suffer from the scarcity of informative training data, leading to overfitting and poor generalization in deep learning models. Data augmentation offers a promising approach to increasing the size and diversity of training data, where faithfulness and diversity are two critical factors to effectiveness. However, promoting untargeted diversity can result in redundant samples with limited utility. To address these challenges, we propose a novel diffusion-based data augmentation approach, Sparse-Aware Semantic-Guided Diffusion Augmentation (SASG-DA). To enhance generation faithfulness, we introduce the Semantic Representation Guidance (SRG) mechanism by leveraging fine-grained, task-aware semantic representations as generation conditions. To enable flexible and diverse sample generation, we propose a Gaussian Modeling Semantic Sampling (GMSS) strategy, which models the semantic representation distribution and allows stochastic sampling to produce both faithful and diverse samples. To enhance targeted diversity, we further introduce a Sparse-Aware Semantic Sampling (SASS) strategy to explicitly explore underrepresented regions, improving distribution coverage and sample utility. Extensive experiments on benchmark sEMG datasets, Ninapro DB2, DB4, and DB7, demonstrate that SASG-DA significantly outperforms existing augmentation methods. Overall, our proposed data augmentation approach effectively mitigates overfitting and improves recognition performance and generalization by offering both faithful and diverse samples.
GNN-ACLP: Graph Neural Networks based Analog Circuit Link Prediction
Apr 14, 2025Abstract:Circuit link prediction identifying missing component connections from incomplete netlists is crucial in automating analog circuit design. However, existing methods face three main challenges: 1) Insufficient use of topological patterns in circuit graphs reduces prediction accuracy; 2) Data scarcity due to the complexity of annotations hinders model generalization; 3) Limited adaptability to various netlist formats. We propose GNN-ACLP, a Graph Neural Networks (GNNs) based framework featuring three innovations to tackle these challenges. First, we introduce the SEAL (Subgraphs, Embeddings, and Attributes for Link Prediction) framework and achieve port-level accuracy in circuit link prediction. Second, we propose Netlist Babel Fish, a netlist format conversion tool leveraging retrieval-augmented generation (RAG) with large language model (LLM) to enhance the compatibility of netlist formats. Finally, we construct SpiceNetlist, a comprehensive dataset that contains 775 annotated circuits across 10 different classes of components. The experimental results demonstrate an improvement of 15.05% on the SpiceNetlist dataset and 12.01% on the Image2Net dataset over the existing approach.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Adaptive Interactive Segmentation for Multimodal Medical Imaging via Selection Engine
Nov 29, 2024Abstract:In medical image analysis, achieving fast, efficient, and accurate segmentation is essential for automated diagnosis and treatment. Although recent advancements in deep learning have significantly improved segmentation accuracy, current models often face challenges in adaptability and generalization, particularly when processing multi-modal medical imaging data. These limitations stem from the substantial variations between imaging modalities and the inherent complexity of medical data. To address these challenges, we propose the Strategy-driven Interactive Segmentation Model (SISeg), built on SAM2, which enhances segmentation performance across various medical imaging modalities by integrating a selection engine. To mitigate memory bottlenecks and optimize prompt frame selection during the inference of 2D image sequences, we developed an automated system, the Adaptive Frame Selection Engine (AFSE). This system dynamically selects the optimal prompt frames without requiring extensive prior medical knowledge and enhances the interpretability of the model's inference process through an interactive feedback mechanism. We conducted extensive experiments on 10 datasets covering 7 representative medical imaging modalities, demonstrating the SISeg model's robust adaptability and generalization in multi-modal tasks. The project page and code will be available at: [URL].
SRSA: A Cost-Efficient Strategy-Router Search Agent for Real-world Human-Machine Interactions
Nov 21, 2024



Abstract:Recently, as Large Language Models (LLMs) have shown impressive emerging capabilities and gained widespread popularity, research on LLM-based search agents has proliferated. In real-world situations, users often input contextual and highly personalized queries to chatbots, challenging LLMs to capture context and generate appropriate answers. However, much of the prior research has not focused specifically on authentic human-machine dialogue scenarios. It also ignores the important balance between response quality and computational cost by forcing all queries to follow the same agent process. To address these gaps, we propose a Strategy-Router Search Agent (SRSA), routing different queries to appropriate search strategies and enabling fine-grained serial searches to obtain high-quality results at a relatively low cost. To evaluate our work, we introduce a new dataset, Contextual Query Enhancement Dataset (CQED), comprising contextual queries to simulate authentic and daily interactions between humans and chatbots. Using LLM-based automatic evaluation metrics, we assessed SRSA's performance in terms of informativeness, completeness, novelty, and actionability. To conclude, SRSA provides an approach that resolves the issue of simple serial searches leading to degenerate answers for lengthy and contextual queries, effectively and efficiently parses complex user queries, and generates more comprehensive and informative responses without fine-tuning an LLM.
Multi-modal Intermediate Feature Interaction AutoEncoder for Overall Survival Prediction of Esophageal Squamous Cell Cancer
Aug 23, 2024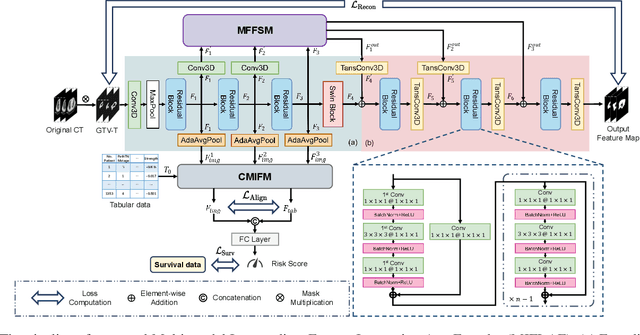
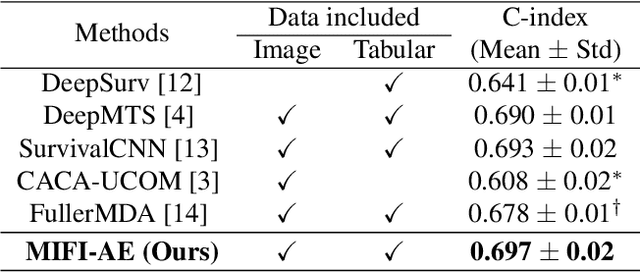
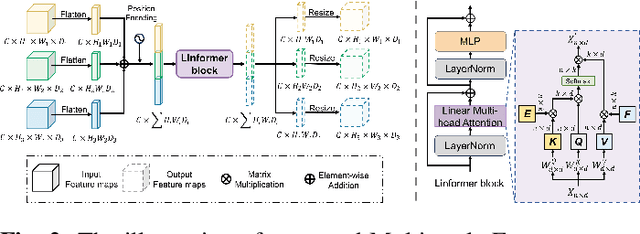
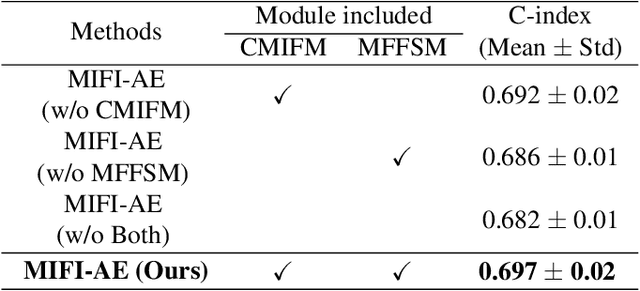
Abstract:Survival prediction for esophageal squamous cell cancer (ESCC) is crucial for doctors to assess a patient's condition and tailor treatment plans. The application and development of multi-modal deep learning in this field have attracted attention in recent years. However, the prognostically relevant features between cross-modalities have not been further explored in previous studies, which could hinder the performance of the model. Furthermore, the inherent semantic gap between different modal feature representations is also ignored. In this work, we propose a novel autoencoder-based deep learning model to predict the overall survival of the ESCC. Two novel modules were designed for multi-modal prognosis-related feature reinforcement and modeling ability enhancement. In addition, a novel joint loss was proposed to make the multi-modal feature representations more aligned. Comparison and ablation experiments demonstrated that our model can achieve satisfactory results in terms of discriminative ability, risk stratification, and the effectiveness of the proposed modules.
Class-balanced Open-set Semi-supervised Object Detection for Medical Images
Aug 22, 2024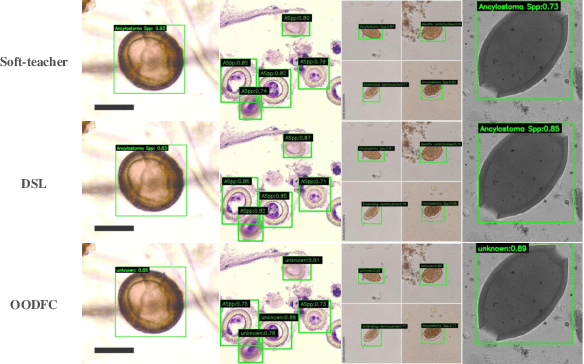
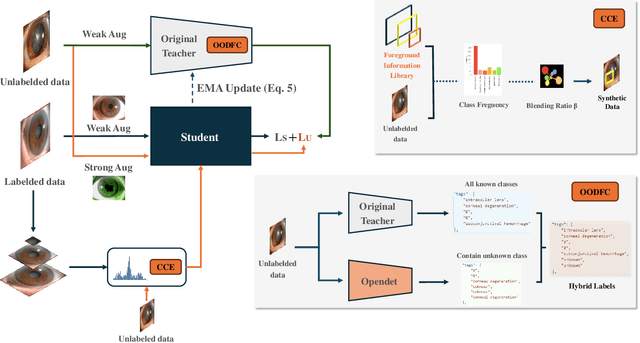
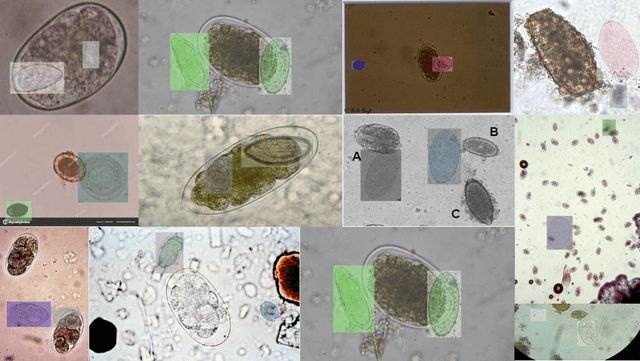
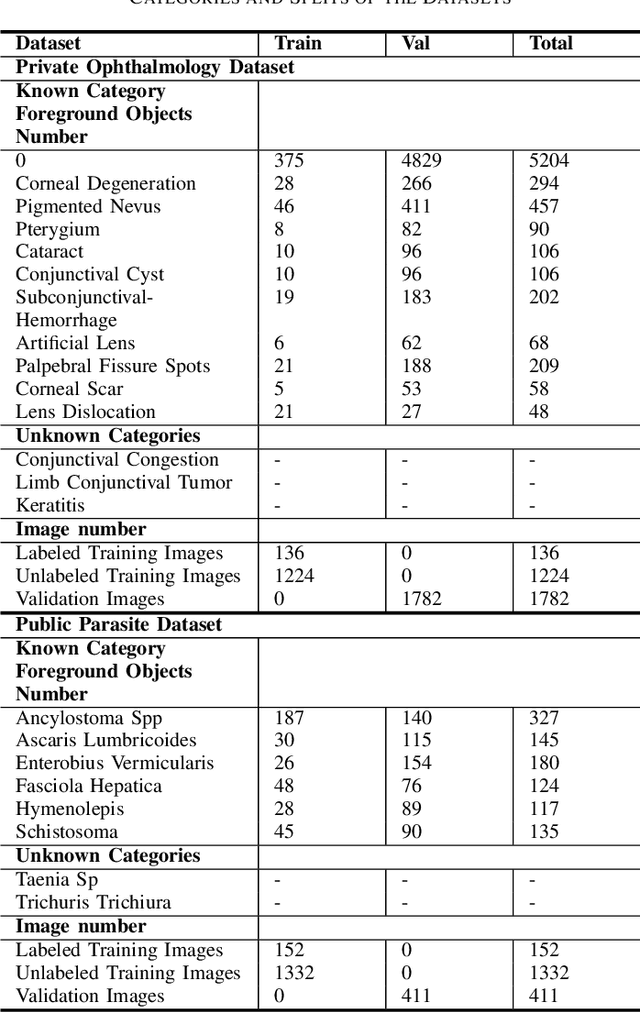
Abstract:Medical image datasets in the real world are often unlabeled and imbalanced, and Semi-Supervised Object Detection (SSOD) can utilize unlabeled data to improve an object detector. However, existing approaches predominantly assumed that the unlabeled data and test data do not contain out-of-distribution (OOD) classes. The few open-set semi-supervised object detection methods have two weaknesses: first, the class imbalance is not considered; second, the OOD instances are distinguished and simply discarded during pseudo-labeling. In this paper, we consider the open-set semi-supervised object detection problem which leverages unlabeled data that contain OOD classes to improve object detection for medical images. Our study incorporates two key innovations: Category Control Embed (CCE) and out-of-distribution Detection Fusion Classifier (OODFC). CCE is designed to tackle dataset imbalance by constructing a Foreground information Library, while OODFC tackles open-set challenges by integrating the ``unknown'' information into basic pseudo-labels. Our method outperforms the state-of-the-art SSOD performance, achieving a 4.25 mAP improvement on the public Parasite dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge