Juan Ye
MAC-Gaze: Motion-Aware Continual Calibration for Mobile Gaze Tracking
May 28, 2025Abstract:Mobile gaze tracking faces a fundamental challenge: maintaining accuracy as users naturally change their postures and device orientations. Traditional calibration approaches, like one-off, fail to adapt to these dynamic conditions, leading to degraded performance over time. We present MAC-Gaze, a Motion-Aware continual Calibration approach that leverages smartphone Inertial measurement unit (IMU) sensors and continual learning techniques to automatically detect changes in user motion states and update the gaze tracking model accordingly. Our system integrates a pre-trained visual gaze estimator and an IMU-based activity recognition model with a clustering-based hybrid decision-making mechanism that triggers recalibration when motion patterns deviate significantly from previously encountered states. To enable accumulative learning of new motion conditions while mitigating catastrophic forgetting, we employ replay-based continual learning, allowing the model to maintain performance across previously encountered motion conditions. We evaluate our system through extensive experiments on the publicly available RGBDGaze dataset and our own 10-hour multimodal MotionGaze dataset (481K+ images, 800K+ IMU readings), encompassing a wide range of postures under various motion conditions including sitting, standing, lying, and walking. Results demonstrate that our method reduces gaze estimation error by 19.9% on RGBDGaze (from 1.73 cm to 1.41 cm) and by 31.7% on MotionGaze (from 2.81 cm to 1.92 cm) compared to traditional calibration approaches. Our framework provides a robust solution for maintaining gaze estimation accuracy in mobile scenarios.
Approximate Maximum Likelihood Inference for Acoustic Spatial Capture-Recapture with Unknown Identities, Using Monte Carlo Expectation Maximization
Oct 06, 2024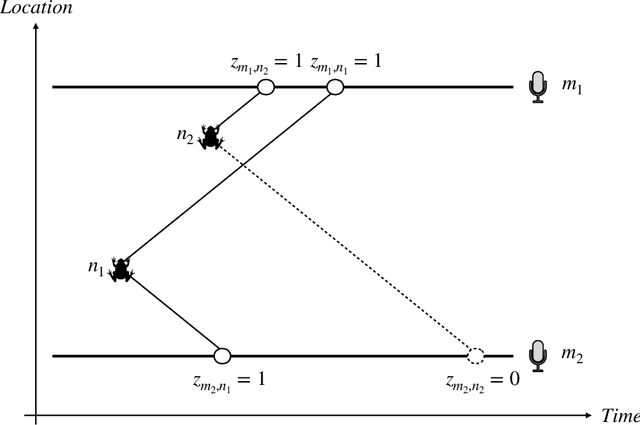
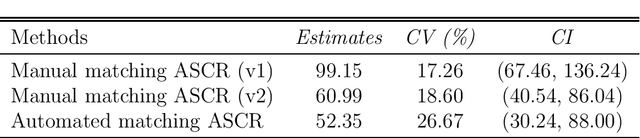
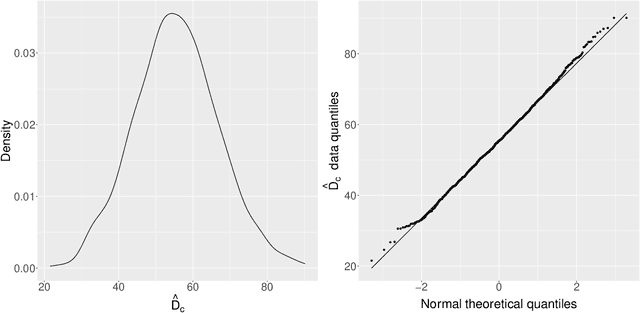

Abstract:Acoustic spatial capture-recapture (ASCR) surveys with an array of synchronized acoustic detectors can be an effective way of estimating animal density or call density. However, constructing the capture histories required for ASCR analysis is challenging, as recognizing which detections at different detectors are of which calls is not a trivial task. Because calls from different distances take different times to arrive at detectors, the order in which calls are detected is not necessarily the same as the order in which they are made, and without knowing which detections are of the same call, we do not know how many different calls are detected. We propose a Monte Carlo expectation-maximization (MCEM) estimation method to resolve this unknown call identity problem. To implement the MCEM method in this context, we sample the latent variables from a complete-data likelihood model in the expectation step and use a semi-complete-data likelihood or conditional likelihood in the maximization step. We use a parametric bootstrap to obtain confidence intervals. When we apply our method to a survey of moss frogs, it gives an estimate within 15% of the estimate obtained using data with call capture histories constructed by experts, and unlike this latter estimate, our confidence interval incorporates the uncertainty about call identities. Simulations show it to have a low bias (6%) and coverage probabilities close to the nominal 95% value.
LPUWF-LDM: Enhanced Latent Diffusion Model for Precise Late-phase UWF-FA Generation on Limited Dataset
Sep 01, 2024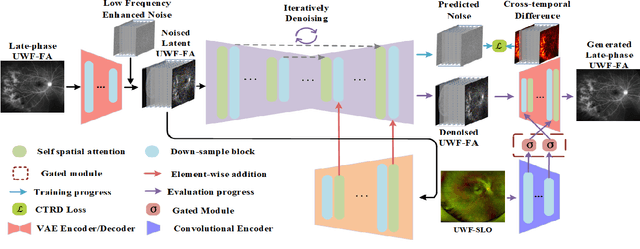
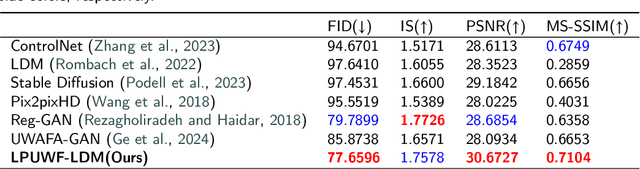
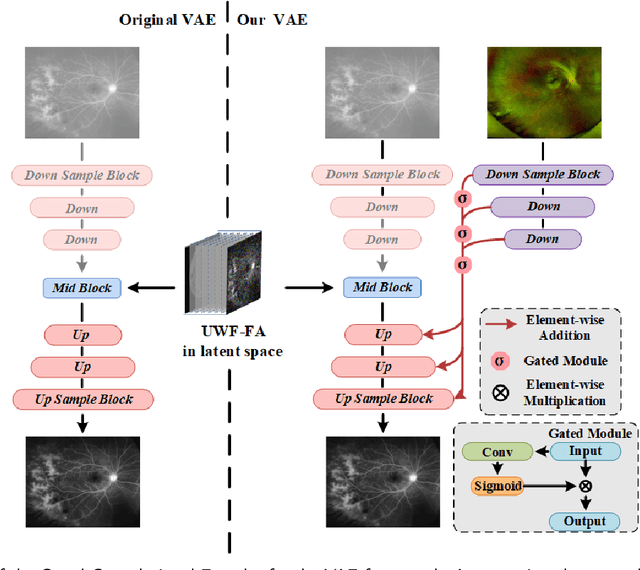

Abstract:Ultra-Wide-Field Fluorescein Angiography (UWF-FA) enables precise identification of ocular diseases using sodium fluorescein, which can be potentially harmful. Existing research has developed methods to generate UWF-FA from Ultra-Wide-Field Scanning Laser Ophthalmoscopy (UWF-SLO) to reduce the adverse reactions associated with injections. However, these methods have been less effective in producing high-quality late-phase UWF-FA, particularly in lesion areas and fine details. Two primary challenges hinder the generation of high-quality late-phase UWF-FA: the scarcity of paired UWF-SLO and early/late-phase UWF-FA datasets, and the need for realistic generation at lesion sites and potential blood leakage regions. This study introduces an improved latent diffusion model framework to generate high-quality late-phase UWF-FA from limited paired UWF images. To address the challenges as mentioned earlier, our approach employs a module utilizing Cross-temporal Regional Difference Loss, which encourages the model to focus on the differences between early and late phases. Additionally, we introduce a low-frequency enhanced noise strategy in the diffusion forward process to improve the realism of medical images. To further enhance the mapping capability of the variational autoencoder module, especially with limited datasets, we implement a Gated Convolutional Encoder to extract additional information from conditional images. Our Latent Diffusion Model for Ultra-Wide-Field Late-Phase Fluorescein Angiography (LPUWF-LDM) effectively reconstructs fine details in late-phase UWF-FA and achieves state-of-the-art results compared to other existing methods when working with limited datasets. Our source code is available at: https://github.com/Tinysqua/****.
Class-balanced Open-set Semi-supervised Object Detection for Medical Images
Aug 22, 2024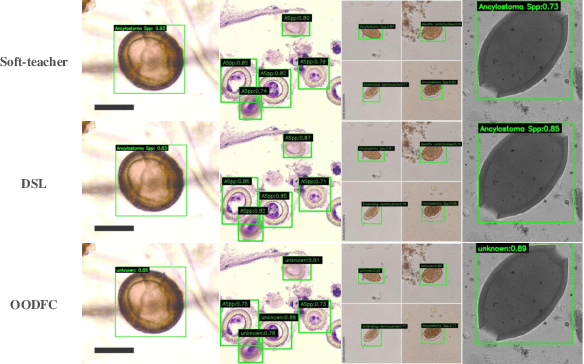
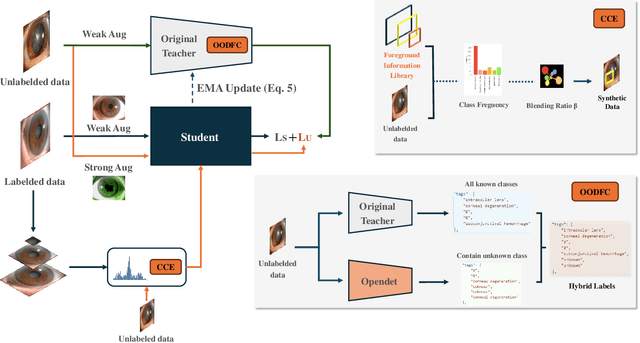
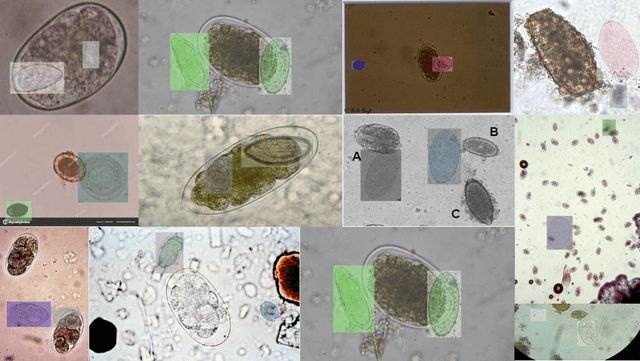
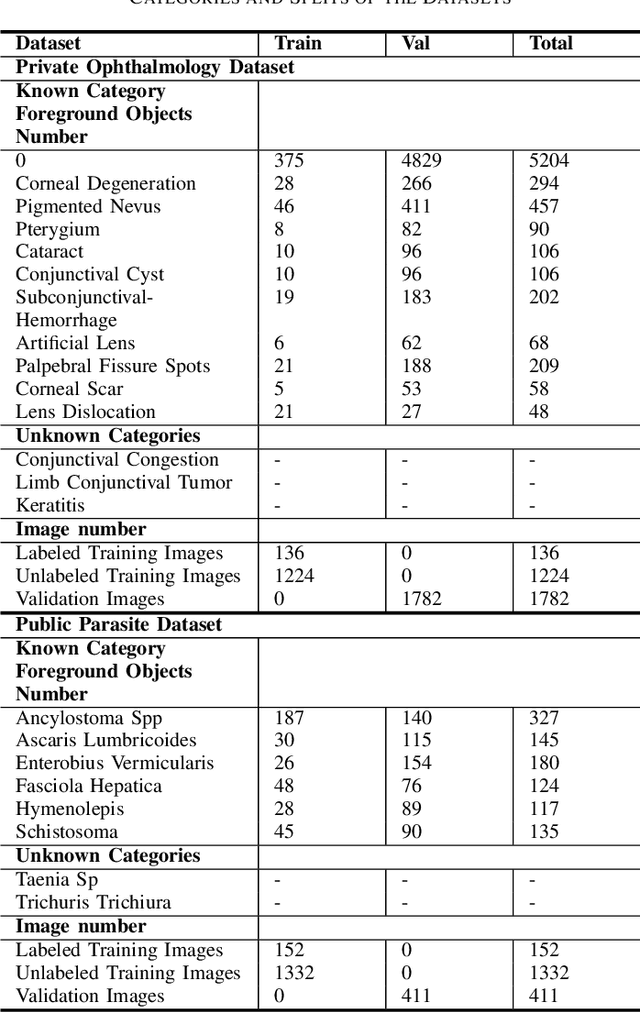
Abstract:Medical image datasets in the real world are often unlabeled and imbalanced, and Semi-Supervised Object Detection (SSOD) can utilize unlabeled data to improve an object detector. However, existing approaches predominantly assumed that the unlabeled data and test data do not contain out-of-distribution (OOD) classes. The few open-set semi-supervised object detection methods have two weaknesses: first, the class imbalance is not considered; second, the OOD instances are distinguished and simply discarded during pseudo-labeling. In this paper, we consider the open-set semi-supervised object detection problem which leverages unlabeled data that contain OOD classes to improve object detection for medical images. Our study incorporates two key innovations: Category Control Embed (CCE) and out-of-distribution Detection Fusion Classifier (OODFC). CCE is designed to tackle dataset imbalance by constructing a Foreground information Library, while OODFC tackles open-set challenges by integrating the ``unknown'' information into basic pseudo-labels. Our method outperforms the state-of-the-art SSOD performance, achieving a 4.25 mAP improvement on the public Parasite dataset.
TTMFN: Two-stream Transformer-based Multimodal Fusion Network for Survival Prediction
Nov 13, 2023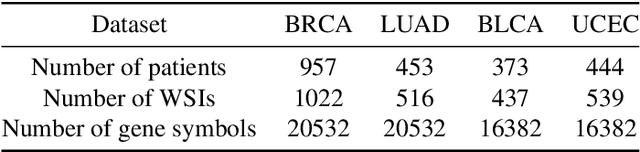
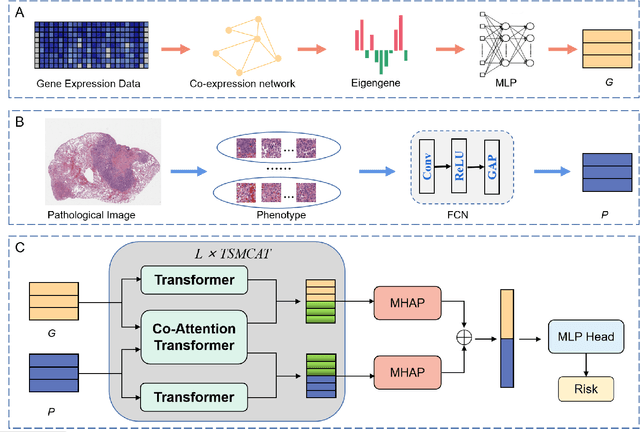
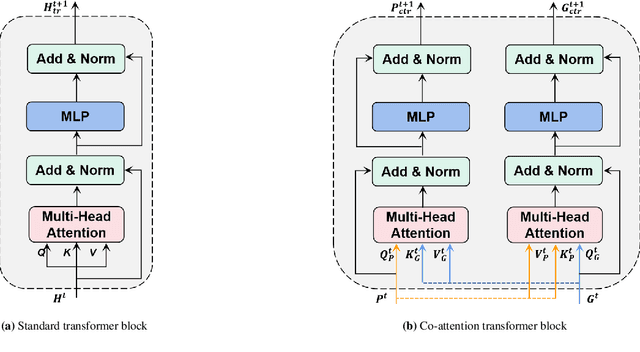
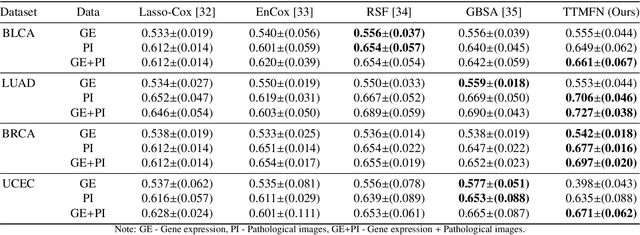
Abstract:Survival prediction plays a crucial role in assisting clinicians with the development of cancer treatment protocols. Recent evidence shows that multimodal data can help in the diagnosis of cancer disease and improve survival prediction. Currently, deep learning-based approaches have experienced increasing success in survival prediction by integrating pathological images and gene expression data. However, most existing approaches overlook the intra-modality latent information and the complex inter-modality correlations. Furthermore, existing modalities do not fully exploit the immense representational capabilities of neural networks for feature aggregation and disregard the importance of relationships between features. Therefore, it is highly recommended to address these issues in order to enhance the prediction performance by proposing a novel deep learning-based method. We propose a novel framework named Two-stream Transformer-based Multimodal Fusion Network for survival prediction (TTMFN), which integrates pathological images and gene expression data. In TTMFN, we present a two-stream multimodal co-attention transformer module to take full advantage of the complex relationships between different modalities and the potential connections within the modalities. Additionally, we develop a multi-head attention pooling approach to effectively aggregate the feature representations of the two modalities. The experiment results on four datasets from The Cancer Genome Atlas demonstrate that TTMFN can achieve the best performance or competitive results compared to the state-of-the-art methods in predicting the overall survival of patients.
Hadamard Domain Training with Integers for Class Incremental Quantized Learning
Oct 05, 2023Abstract:Continual learning is a desirable feature in many modern machine learning applications, which allows in-field adaptation and updating, ranging from accommodating distribution shift, to fine-tuning, and to learning new tasks. For applications with privacy and low latency requirements, the compute and memory demands imposed by continual learning can be cost-prohibitive for resource-constraint edge platforms. Reducing computational precision through fully quantized training (FQT) simultaneously reduces memory footprint and increases compute efficiency for both training and inference. However, aggressive quantization especially integer FQT typically degrades model accuracy to unacceptable levels. In this paper, we propose a technique that leverages inexpensive Hadamard transforms to enable low-precision training with only integer matrix multiplications. We further determine which tensors need stochastic rounding and propose tiled matrix multiplication to enable low-bit width accumulators. We demonstrate the effectiveness of our technique on several human activity recognition datasets and CIFAR100 in a class incremental learning setting. We achieve less than 0.5% and 3% accuracy degradation while we quantize all matrix multiplications inputs down to 4-bits with 8-bit accumulators.
Towards Automated Animal Density Estimation with Acoustic Spatial Capture-Recapture
Aug 24, 2023



Abstract:Passive acoustic monitoring can be an effective way of monitoring wildlife populations that are acoustically active but difficult to survey visually. Digital recorders allow surveyors to gather large volumes of data at low cost, but identifying target species vocalisations in these data is non-trivial. Machine learning (ML) methods are often used to do the identification. They can process large volumes of data quickly, but they do not detect all vocalisations and they do generate some false positives (vocalisations that are not from the target species). Existing wildlife abundance survey methods have been designed specifically to deal with the first of these mistakes, but current methods of dealing with false positives are not well-developed. They do not take account of features of individual vocalisations, some of which are more likely to be false positives than others. We propose three methods for acoustic spatial capture-recapture inference that integrate individual-level measures of confidence from ML vocalisation identification into the likelihood and hence integrate ML uncertainty into inference. The methods include a mixture model in which species identity is a latent variable. We test the methods by simulation and find that in a scenario based on acoustic data from Hainan gibbons, in which ignoring false positives results in 17% positive bias, our methods give negligible bias and coverage probabilities that are close to the nominal 95% level.
An End-to-End Review of Gaze Estimation and its Interactive Applications on Handheld Mobile Devices
Jun 30, 2023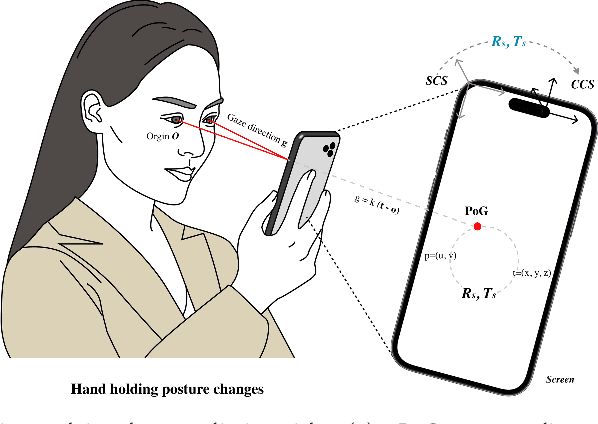
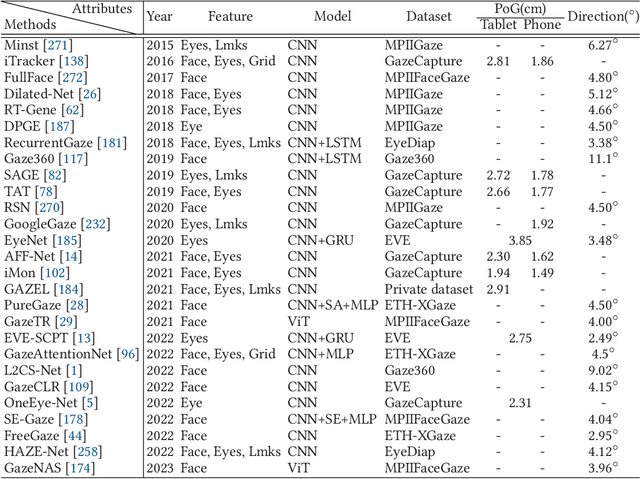
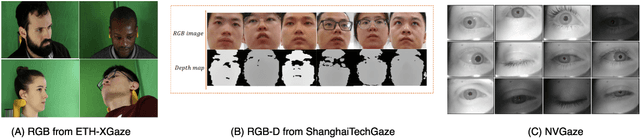
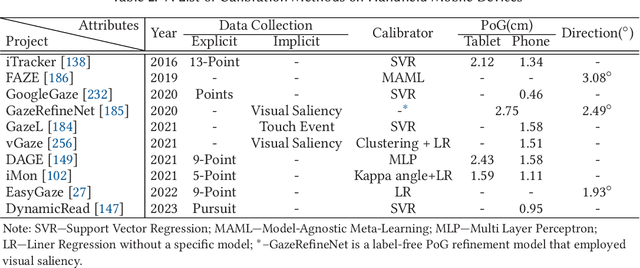
Abstract:In recent years we have witnessed an increasing number of interactive systems on handheld mobile devices which utilise gaze as a single or complementary interaction modality. This trend is driven by the enhanced computational power of these devices, higher resolution and capacity of their cameras, and improved gaze estimation accuracy obtained from advanced machine learning techniques, especially in deep learning. As the literature is fast progressing, there is a pressing need to review the state of the art, delineate the boundary, and identify the key research challenges and opportunities in gaze estimation and interaction. This paper aims to serve this purpose by presenting an end-to-end holistic view in this area, from gaze capturing sensors, to gaze estimation workflows, to deep learning techniques, and to gaze interactive applications.
* 37 Pages, Paper accepted by ACM Computing Surveys
A simple normalization technique using window statistics to improve the out-of-distribution generalization on medical images
Jul 14, 2022



Abstract:Since data scarcity and data heterogeneity are prevailing for medical images, well-trained Convolutional Neural Networks (CNNs) using previous normalization methods may perform poorly when deployed to a new site. However, a reliable model for real-world clinical applications should be able to generalize well both on in-distribution (IND) and out-of-distribution (OOD) data (e.g., the new site data). In this study, we present a novel normalization technique called window normalization (WIN) to improve the model generalization on heterogeneous medical images, which is a simple yet effective alternative to existing normalization methods. Specifically, WIN perturbs the normalizing statistics with the local statistics computed on the window of features. This feature-level augmentation technique regularizes the models well and improves their OOD generalization significantly. Taking its advantage, we propose a novel self-distillation method called WIN-WIN for classification tasks. WIN-WIN is easily implemented with twice forward passes and a consistency constraint, which can be a simple extension for existing methods. Extensive experimental results on various tasks (6 tasks) and datasets (24 datasets) demonstrate the generality and effectiveness of our methods.
CDNet: Contrastive Disentangled Network for Fine-Grained Image Categorization of Ocular B-Scan Ultrasound
Jun 17, 2022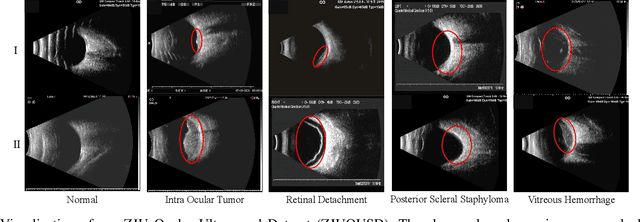
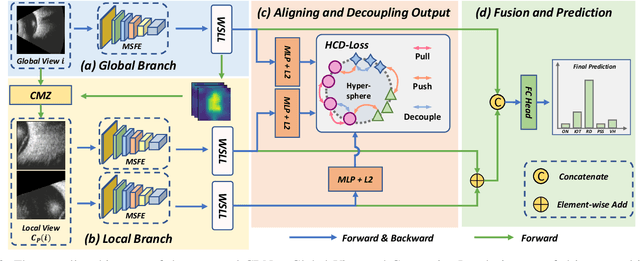
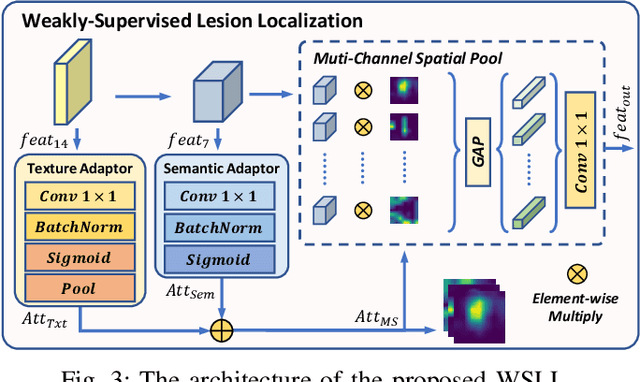
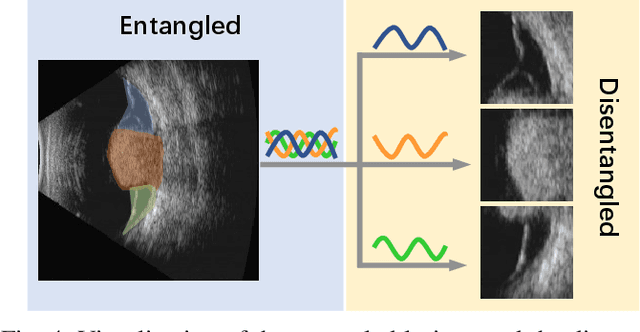
Abstract:Precise and rapid categorization of images in the B-scan ultrasound modality is vital for diagnosing ocular diseases. Nevertheless, distinguishing various diseases in ultrasound still challenges experienced ophthalmologists. Thus a novel contrastive disentangled network (CDNet) is developed in this work, aiming to tackle the fine-grained image categorization (FGIC) challenges of ocular abnormalities in ultrasound images, including intraocular tumor (IOT), retinal detachment (RD), posterior scleral staphyloma (PSS), and vitreous hemorrhage (VH). Three essential components of CDNet are the weakly-supervised lesion localization module (WSLL), contrastive multi-zoom (CMZ) strategy, and hyperspherical contrastive disentangled loss (HCD-Loss), respectively. These components facilitate feature disentanglement for fine-grained recognition in both the input and output aspects. The proposed CDNet is validated on our ZJU Ocular Ultrasound Dataset (ZJUOUSD), consisting of 5213 samples. Furthermore, the generalization ability of CDNet is validated on two public and widely-used chest X-ray FGIC benchmarks. Quantitative and qualitative results demonstrate the efficacy of our proposed CDNet, which achieves state-of-the-art performance in the FGIC task. Code is available at: https://github.com/ZeroOneGame/CDNet-for-OUS-FGIC .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge