Ruilong Dan
ChatDoctor: A Medical Chat Model Fine-tuned on LLaMA Model using Medical Domain Knowledge
Apr 01, 2023Abstract:Recent large language models (LLMs) in the general domain, such as ChatGPT, have shown remarkable success in following instructions and producing human-like responses. However, such language models have not been tailored to the medical domain, resulting in poor answer accuracy and inability to give plausible recommendations for medical diagnosis, medications, etc. To address this issue, we collected more than 700 diseases and their corresponding symptoms, required medical tests, and recommended medications, from which we generated 5K doctor-patient conversations. In addition, we obtained 200K real patient-doctor conversations from online Q\&A medical consultation sites. By fine-tuning LLMs using these 205k doctor-patient conversations, the resulting models emerge with great potential to understand patients' needs, provide informed advice, and offer valuable assistance in a variety of medical-related fields. The integration of these advanced language models into healthcare can revolutionize the way healthcare professionals and patients communicate, ultimately improving the overall efficiency and quality of patient care and outcomes. In addition, we made public all the source codes, datasets, and model weights to facilitate the further development of dialogue models in the medical field. The training data, codes, and weights of this project are available at: The training data, codes, and weights of this project are available at: https://github.com/Kent0n-Li/ChatDoctor.
CDNet: Contrastive Disentangled Network for Fine-Grained Image Categorization of Ocular B-Scan Ultrasound
Jun 17, 2022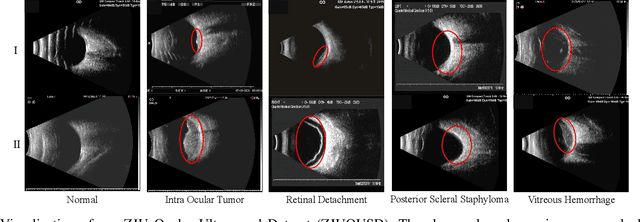
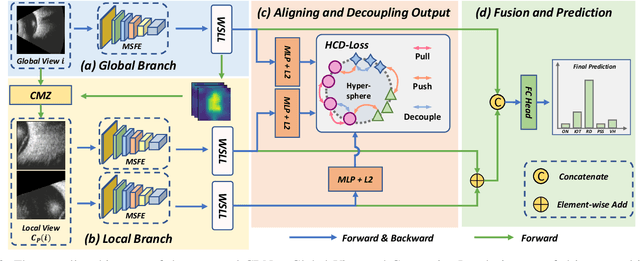
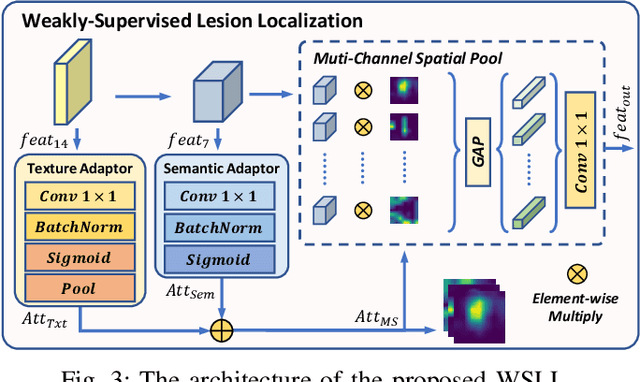
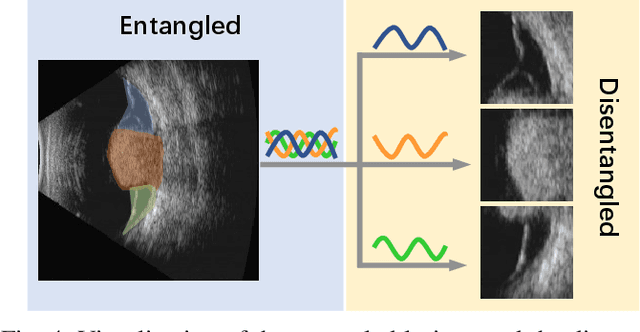
Abstract:Precise and rapid categorization of images in the B-scan ultrasound modality is vital for diagnosing ocular diseases. Nevertheless, distinguishing various diseases in ultrasound still challenges experienced ophthalmologists. Thus a novel contrastive disentangled network (CDNet) is developed in this work, aiming to tackle the fine-grained image categorization (FGIC) challenges of ocular abnormalities in ultrasound images, including intraocular tumor (IOT), retinal detachment (RD), posterior scleral staphyloma (PSS), and vitreous hemorrhage (VH). Three essential components of CDNet are the weakly-supervised lesion localization module (WSLL), contrastive multi-zoom (CMZ) strategy, and hyperspherical contrastive disentangled loss (HCD-Loss), respectively. These components facilitate feature disentanglement for fine-grained recognition in both the input and output aspects. The proposed CDNet is validated on our ZJU Ocular Ultrasound Dataset (ZJUOUSD), consisting of 5213 samples. Furthermore, the generalization ability of CDNet is validated on two public and widely-used chest X-ray FGIC benchmarks. Quantitative and qualitative results demonstrate the efficacy of our proposed CDNet, which achieves state-of-the-art performance in the FGIC task. Code is available at: https://github.com/ZeroOneGame/CDNet-for-OUS-FGIC .
Source-free Domain Adaptation for Multi-site and Lifespan Brain Skull Stripping
Mar 11, 2022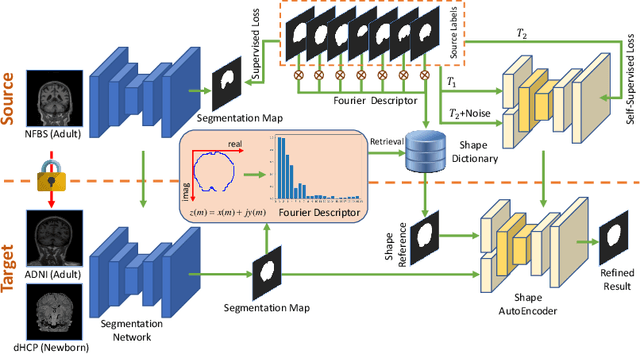
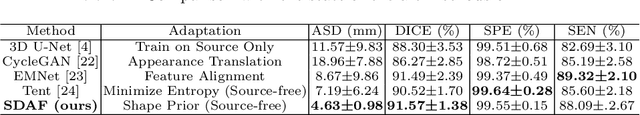
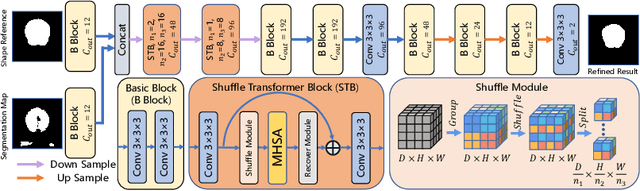
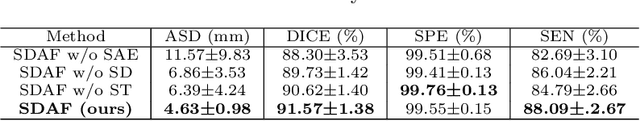
Abstract:Skull stripping is a crucial prerequisite step in the analysis of brain magnetic resonance (MR) images. Although many excellent works or tools have been proposed, they suffer from low generalization capability. For instance, the model trained on a dataset with specific imaging parameters (source domain) cannot be well applied to other datasets with different imaging parameters (target domain). Especially, for the lifespan datasets, the model trained on an adult dataset is not applicable to an infant dataset due to the large domain difference. To address this issue, numerous domain adaptation (DA) methods have been proposed to align the extracted features between the source and target domains, requiring concurrent access to the input images of both domains. Unfortunately, it is problematic to share the images due to privacy. In this paper, we design a source-free domain adaptation framework (SDAF) for multi-site and lifespan skull stripping that can accomplish domain adaptation without access to source domain images. Our method only needs to share the source labels as shape dictionaries and the weights trained on the source data, without disclosing private information from source domain subjects. To deal with the domain shift between multi-site lifespan datasets, we take advantage of the brain shape prior which is invariant to imaging parameters and ages. Experiments demonstrate that our framework can significantly outperform the state-of-the-art methods on multi-site lifespan datasets.
Dispensed Transformer Network for Unsupervised Domain Adaptation
Oct 28, 2021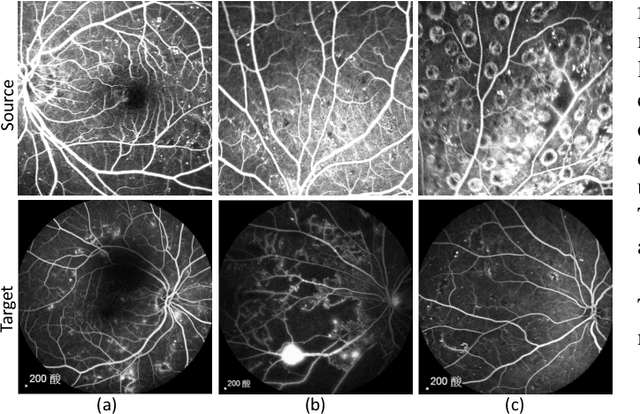
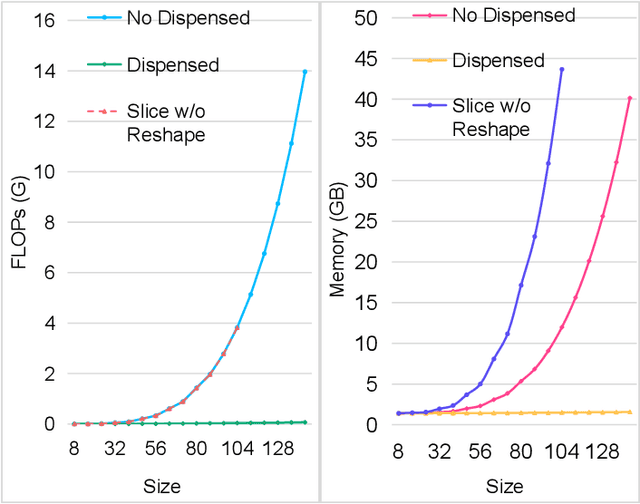
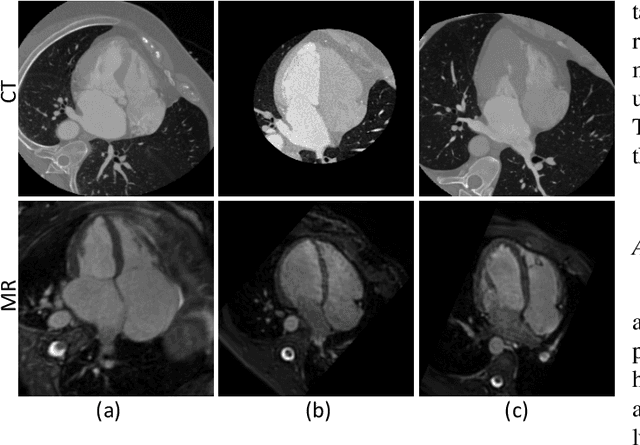
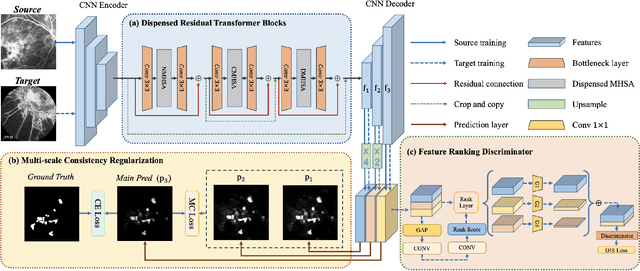
Abstract:Accurate segmentation is a crucial step in medical image analysis and applying supervised machine learning to segment the organs or lesions has been substantiated effective. However, it is costly to perform data annotation that provides ground truth labels for training the supervised algorithms, and the high variance of data that comes from different domains tends to severely degrade system performance over cross-site or cross-modality datasets. To mitigate this problem, a novel unsupervised domain adaptation (UDA) method named dispensed Transformer network (DTNet) is introduced in this paper. Our novel DTNet contains three modules. First, a dispensed residual transformer block is designed, which realizes global attention by dispensed interleaving operation and deals with the excessive computational cost and GPU memory usage of the Transformer. Second, a multi-scale consistency regularization is proposed to alleviate the loss of details in the low-resolution output for better feature alignment. Finally, a feature ranking discriminator is introduced to automatically assign different weights to domain-gap features to lessen the feature distribution distance, reducing the performance shift of two domains. The proposed method is evaluated on large fluorescein angiography (FA) retinal nonperfusion (RNP) cross-site dataset with 676 images and a wide used cross-modality dataset from the MM-WHS challenge. Extensive results demonstrate that our proposed network achieves the best performance in comparison with several state-of-the-art techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge