Yifei Chen
Just-in-Time Catching Test Generation at Meta
Jan 30, 2026Abstract:We report on Just-in-Time catching test generation at Meta, designed to prevent bugs in large scale backend systems of hundreds of millions of line of code. Unlike traditional hardening tests, which pass at generation time, catching tests are meant to fail, surfacing bugs before code lands. The primary challenge is to reduce development drag from false positive test failures. Analyzing 22,126 generated tests, we show code-change-aware methods improve candidate catch generation 4x over hardening tests and 20x over coincidentally failing tests. To address false positives, we use rule-based and LLM-based assessors. These assessors reduce human review load by 70%. Inferential statistical analysis showed that human-accepted code changes are assessed to have significantly more false positives, while human-rejected changes have significantly more true positives. We reported 41 candidate catches to engineers; 8 were confirmed to be true positives, 4 of which would have led to serious failures had they remained uncaught. Overall, our results show that Just-in-Time catching is scalable, industrially applicable, and that it prevents serious failures from reaching production.
Forward Consistency Learning with Gated Context Aggregation for Video Anomaly Detection
Jan 26, 2026Abstract:As a crucial element of public security, video anomaly detection (VAD) aims to measure deviations from normal patterns for various events in real-time surveillance systems. However, most existing VAD methods rely on large-scale models to pursue extreme accuracy, limiting their feasibility on resource-limited edge devices. Moreover, mainstream prediction-based VAD detects anomalies using only single-frame future prediction errors, overlooking the richer constraints from longer-term temporal forward information. In this paper, we introduce FoGA, a lightweight VAD model that performs Forward consistency learning with Gated context Aggregation, containing about 2M parameters and tailored for potential edge devices. Specifically, we propose a Unet-based method that performs feature extraction on consecutive frames to generate both immediate and forward predictions. Then, we introduce a gated context aggregation module into the skip connections to dynamically fuse encoder and decoder features at the same spatial scale. Finally, the model is jointly optimized with a novel forward consistency loss, and a hybrid anomaly measurement strategy is adopted to integrate errors from both immediate and forward frames for more accurate detection. Extensive experiments demonstrate the effectiveness of the proposed method, which substantially outperforms state-of-the-art competing methods, running up to 155 FPS. Hence, our FoGA achieves an excellent trade-off between performance and the efficiency metric.
SMc2f: Robust Scenario Mining for Robotic Autonomy from Coarse to Fine
Jan 17, 2026Abstract:The safety validation of autonomous robotic vehicles hinges on systematically testing their planning and control stacks against rare, safety-critical scenarios. Mining these long-tail events from massive real-world driving logs is therefore a critical step in the robotic development lifecycle. The goal of the Scenario Mining task is to retrieve useful information to enable targeted re-simulation, regression testing, and failure analysis of the robot's decision-making algorithms. RefAV, introduced by the Argoverse team, is an end-to-end framework that uses large language models (LLMs) to spatially and temporally localize scenarios described in natural language. However, this process performs retrieval on trajectory labels, ignoring the direct connection between natural language and raw RGB images, which runs counter to the intuition of video retrieval; it also depends on the quality of upstream 3D object detection and tracking. Further, inaccuracies in trajectory data lead to inaccuracies in downstream spatial and temporal localization. To address these issues, we propose Robust Scenario Mining for Robotic Autonomy from Coarse to Fine (SMc2f), a coarse-to-fine pipeline that employs vision-language models (VLMs) for coarse image-text filtering, builds a database of successful mining cases on top of RefAV and automatically retrieves exemplars to few-shot condition the LLM for more robust retrieval, and introduces text-trajectory contrastive learning to pull matched pairs together and push mismatched pairs apart in a shared embedding space, yielding a fine-grained matcher that refines the LLM's candidate trajectories. Experiments on public datasets demonstrate substantial gains in both retrieval quality and efficiency.
ET-Agent: Incentivizing Effective Tool-Integrated Reasoning Agent via Behavior Calibration
Jan 11, 2026Abstract:Large Language Models (LLMs) can extend their parameter knowledge limits by adopting the Tool-Integrated Reasoning (TIR) paradigm. However, existing LLM-based agent training framework often focuses on answers' accuracy, overlooking specific alignment for behavior patterns. Consequently, agent often exhibits ineffective actions during TIR tasks, such as redundant and insufficient tool calls. How to calibrate erroneous behavioral patterns when executing TIR tasks, thereby exploring effective trajectories, remains an open-ended problem. In this paper, we propose ET-Agent, a training framework for calibrating agent's tool-use behavior through two synergistic perspectives: Self-evolving Data Flywheel and Behavior Calibration Training. Specifically, we introduce a self-evolutionary data flywheel to generate enhanced data, used to fine-tune LLM to improve its exploration ability. Based on this, we implement an two-phases behavior-calibration training framework. It is designed to progressively calibrate erroneous behavioral patterns to optimal behaviors. Further in-depth experiments confirm the superiority of \ourmodel{} across multiple dimensions, including correctness, efficiency, reasoning conciseness, and tool execution accuracy. Our ET-Agent framework provides practical insights for research in the TIR field. Codes can be found in https://github.com/asilverlight/ET-Agent
AnyAD: Unified Any-Modality Anomaly Detection in Incomplete Multi-Sequence MRI
Dec 24, 2025Abstract:Reliable anomaly detection in brain MRI remains challenging due to the scarcity of annotated abnormal cases and the frequent absence of key imaging modalities in real clinical workflows. Existing single-class or multi-class anomaly detection (AD) models typically rely on fixed modality configurations, require repetitive training, or fail to generalize to unseen modality combinations, limiting their clinical scalability. In this work, we present a unified Any-Modality AD framework that performs robust anomaly detection and localization under arbitrary MRI modality availability. The framework integrates a dual-pathway DINOv2 encoder with a feature distribution alignment mechanism that statistically aligns incomplete-modality features with full-modality representations, enabling stable inference even with severe modality dropout. To further enhance semantic consistency, we introduce an Intrinsic Normal Prototypes (INPs) extractor and an INP-guided decoder that reconstruct only normal anatomical patterns while naturally amplifying abnormal deviations. Through randomized modality masking and indirect feature completion during training, the model learns to adapt to all modality configurations without re-training. Extensive experiments on BraTS2018, MU-Glioma-Post, and Pretreat-MetsToBrain-Masks demonstrate that our approach consistently surpasses state-of-the-art industrial and medical AD baselines across 7 modality combinations, achieving superior generalization. This study establishes a scalable paradigm for multimodal medical AD under real-world, imperfect modality conditions. Our source code is available at https://github.com/wuchangw/AnyAD.
Poster: Recognizing Hidden-in-the-Ear Private Key for Reliable Silent Speech Interface Using Multi-Task Learning
Dec 18, 2025Abstract:Silent speech interface (SSI) enables hands-free input without audible vocalization, but most SSI systems do not verify speaker identity. We present HEar-ID, which uses consumer active noise-canceling earbuds to capture low-frequency "whisper" audio and high-frequency ultrasonic reflections. Features from both streams pass through a shared encoder, producing embeddings that feed a contrastive branch for user authentication and an SSI head for silent spelling recognition. This design supports decoding of 50 words while reliably rejecting impostors, all on commodity earbuds with a single model. Experiments demonstrate that HEar-ID achieves strong spelling accuracy and robust authentication.
An Iteration-Free Fixed-Point Estimator for Diffusion Inversion
Dec 09, 2025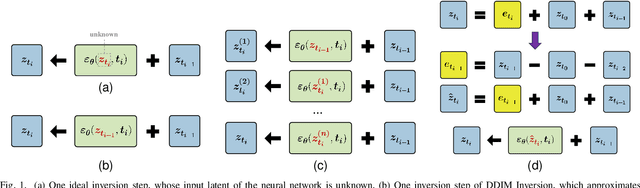


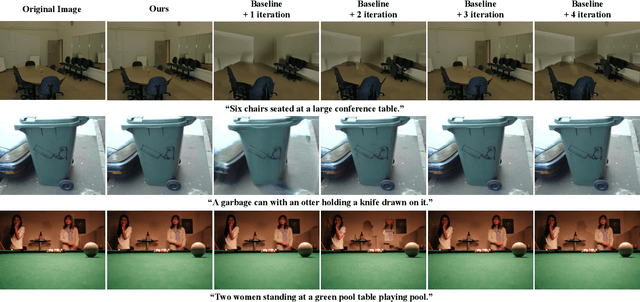
Abstract:Diffusion inversion aims to recover the initial noise corresponding to a given image such that this noise can reconstruct the original image through the denoising diffusion process. The key component of diffusion inversion is to minimize errors at each inversion step, thereby mitigating cumulative inaccuracies. Recently, fixed-point iteration has emerged as a widely adopted approach to minimize reconstruction errors at each inversion step. However, it suffers from high computational costs due to its iterative nature and the complexity of hyperparameter selection. To address these issues, we propose an iteration-free fixed-point estimator for diffusion inversion. First, we derive an explicit expression of the fixed point from an ideal inversion step. Unfortunately, it inherently contains an unknown data prediction error. Building upon this, we introduce the error approximation, which uses the calculable error from the previous inversion step to approximate the unknown error at the current inversion step. This yields a calculable, approximate expression for the fixed point, which is an unbiased estimator characterized by low variance, as shown by our theoretical analysis. We evaluate reconstruction performance on two text-image datasets, NOCAPS and MS-COCO. Compared to DDIM inversion and other inversion methods based on the fixed-point iteration, our method achieves consistent and superior performance in reconstruction tasks without additional iterations or training.
FDP: A Frequency-Decomposition Preprocessing Pipeline for Unsupervised Anomaly Detection in Brain MRI
Nov 17, 2025Abstract:Due to the diversity of brain anatomy and the scarcity of annotated data, supervised anomaly detection for brain MRI remains challenging, driving the development of unsupervised anomaly detection (UAD) approaches. Current UAD methods typically utilize artificially generated noise perturbations on healthy MRIs to train generative models for normal anatomy reconstruction, enabling anomaly detection via residual mapping. However, such simulated anomalies lack the biophysical fidelity and morphological complexity characteristic of true clinical lesions. To advance UAD in brain MRI, we conduct the first systematic frequency-domain analysis of pathological signatures, revealing two key properties: (1) anomalies exhibit unique frequency patterns distinguishable from normal anatomy, and (2) low-frequency signals maintain consistent representations across healthy scans. These insights motivate our Frequency-Decomposition Preprocessing (FDP) framework, the first UAD method to leverage frequency-domain reconstruction for simultaneous pathology suppression and anatomical preservation. FDP can integrate seamlessly with existing anomaly simulation techniques, consistently enhancing detection performance across diverse architectures while maintaining diagnostic fidelity. Experimental results demonstrate that FDP consistently improves anomaly detection performance when integrated with existing methods. Notably, FDP achieves a 17.63% increase in DICE score with LDM while maintaining robust improvements across multiple baselines. The code is available at https://github.com/ls1rius/MRI_FDP.
No Modality Left Behind: Adapting to Missing Modalities via Knowledge Distillation for Brain Tumor Segmentation
Sep 18, 2025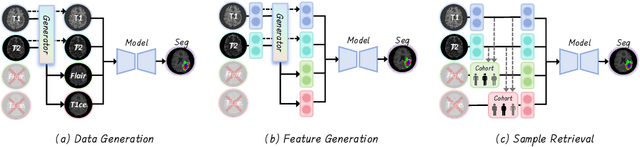
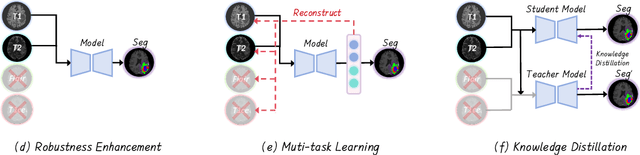
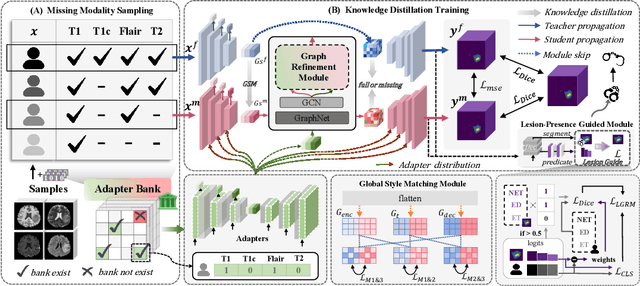
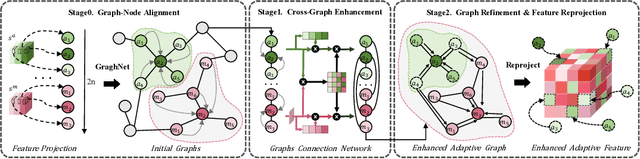
Abstract:Accurate brain tumor segmentation is essential for preoperative evaluation and personalized treatment. Multi-modal MRI is widely used due to its ability to capture complementary tumor features across different sequences. However, in clinical practice, missing modalities are common, limiting the robustness and generalizability of existing deep learning methods that rely on complete inputs, especially under non-dominant modality combinations. To address this, we propose AdaMM, a multi-modal brain tumor segmentation framework tailored for missing-modality scenarios, centered on knowledge distillation and composed of three synergistic modules. The Graph-guided Adaptive Refinement Module explicitly models semantic associations between generalizable and modality-specific features, enhancing adaptability to modality absence. The Bi-Bottleneck Distillation Module transfers structural and textural knowledge from teacher to student models via global style matching and adversarial feature alignment. The Lesion-Presence-Guided Reliability Module predicts prior probabilities of lesion types through an auxiliary classification task, effectively suppressing false positives under incomplete inputs. Extensive experiments on the BraTS 2018 and 2024 datasets demonstrate that AdaMM consistently outperforms existing methods, exhibiting superior segmentation accuracy and robustness, particularly in single-modality and weak-modality configurations. In addition, we conduct a systematic evaluation of six categories of missing-modality strategies, confirming the superiority of knowledge distillation and offering practical guidance for method selection and future research. Our source code is available at https://github.com/Quanato607/AdaMM.
Bridging the Gap in Missing Modalities: Leveraging Knowledge Distillation and Style Matching for Brain Tumor Segmentation
Jul 30, 2025Abstract:Accurate and reliable brain tumor segmentation, particularly when dealing with missing modalities, remains a critical challenge in medical image analysis. Previous studies have not fully resolved the challenges of tumor boundary segmentation insensitivity and feature transfer in the absence of key imaging modalities. In this study, we introduce MST-KDNet, aimed at addressing these critical issues. Our model features Multi-Scale Transformer Knowledge Distillation to effectively capture attention weights at various resolutions, Dual-Mode Logit Distillation to improve the transfer of knowledge, and a Global Style Matching Module that integrates feature matching with adversarial learning. Comprehensive experiments conducted on the BraTS and FeTS 2024 datasets demonstrate that MST-KDNet surpasses current leading methods in both Dice and HD95 scores, particularly in conditions with substantial modality loss. Our approach shows exceptional robustness and generalization potential, making it a promising candidate for real-world clinical applications. Our source code is available at https://github.com/Quanato607/MST-KDNet.
* 11 pages, 2 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge