Dahong Qian
Fusion of Spatio-Temporal and Multi-Scale Frequency Features for Dry Electrodes MI-EEG Decoding
Jan 26, 2026Abstract:Dry-electrode Motor Imagery Electroencephalography (MI-EEG) enables fast, comfortable, real-world Brain Computer Interface by eliminating gels and shortening setup for at-home and wearable use.However, dry recordings pose three main issues: lower Signal-to-Noise Ratio with more baseline drift and sudden transients; weaker and noisier data with poor phase alignment across trials; and bigger variances between sessions. These drawbacks lead to larger data distribution shift, making features less stable for MI-EEG tasks.To address these problems, we introduce STGMFM, a tri-branch framework tailored for dry-electrode MI-EEG, which models complementary spatio-temporal dependencies via dual graph orders, and captures robust envelope dynamics with a multi-scale frequency mixing branch, motivated by the observation that amplitude envelopes are less sensitive to contact variability than instantaneous waveforms. Physiologically meaningful connectivity priors guide learning, and decision-level fusion consolidates a noise-tolerant consensus. On our collected dry-electrode MI-EEG, STGMFM consistently surpasses competitive CNN/Transformer/graph baselines. Codes are available at https://github.com/Tianyi-325/STGMFM.
GPF-Net: Gated Progressive Fusion Learning for Polyp Re-Identification
Dec 25, 2025Abstract:Colonoscopic Polyp Re-Identification aims to match the same polyp from a large gallery with images from different views taken using different cameras, which plays an important role in the prevention and treatment of colorectal cancer in computer-aided diagnosis. However, the coarse resolution of high-level features of a specific polyp often leads to inferior results for small objects where detailed information is important. To address this challenge, we propose a novel architecture, named Gated Progressive Fusion network, to selectively fuse features from multiple levels using gates in a fully connected way for polyp ReID. On the basis of it, a gated progressive fusion strategy is introduced to achieve layer-wise refinement of semantic information through multi-level feature interactions. Experiments on standard benchmarks show the benefits of the multimodal setting over state-of-the-art unimodal ReID models, especially when combined with the specialized multimodal fusion strategy.
LDP: Parameter-Efficient Fine-Tuning of Multimodal LLM for Medical Report Generation
Dec 11, 2025Abstract:Colonoscopic polyp diagnosis is pivotal for early colorectal cancer detection, yet traditional automated reporting suffers from inconsistencies and hallucinations due to the scarcity of high-quality multimodal medical data. To bridge this gap, we propose LDP, a novel framework leveraging multimodal large language models (MLLMs) for professional polyp diagnosis report generation. Specifically, we curate MMEndo, a multimodal endoscopic dataset comprising expert-annotated colonoscopy image-text pairs. We fine-tune the Qwen2-VL-7B backbone using Parameter-Efficient Fine-Tuning (LoRA) and align it with clinical standards via Direct Preference Optimization (DPO). Extensive experiments show that our LDP outperforms existing baselines on both automated metrics and rigorous clinical expert evaluations (achieving a Physician Score of 7.2/10), significantly reducing training computational costs by 833x compared to full fine-tuning. The proposed solution offers a scalable, clinically viable path for primary healthcare, with additional validation on the IU-XRay dataset confirming its robustness.
SASG-DA: Sparse-Aware Semantic-Guided Diffusion Augmentation For Myoelectric Gesture Recognition
Nov 12, 2025Abstract:Surface electromyography (sEMG)-based gesture recognition plays a critical role in human-machine interaction (HMI), particularly for rehabilitation and prosthetic control. However, sEMG-based systems often suffer from the scarcity of informative training data, leading to overfitting and poor generalization in deep learning models. Data augmentation offers a promising approach to increasing the size and diversity of training data, where faithfulness and diversity are two critical factors to effectiveness. However, promoting untargeted diversity can result in redundant samples with limited utility. To address these challenges, we propose a novel diffusion-based data augmentation approach, Sparse-Aware Semantic-Guided Diffusion Augmentation (SASG-DA). To enhance generation faithfulness, we introduce the Semantic Representation Guidance (SRG) mechanism by leveraging fine-grained, task-aware semantic representations as generation conditions. To enable flexible and diverse sample generation, we propose a Gaussian Modeling Semantic Sampling (GMSS) strategy, which models the semantic representation distribution and allows stochastic sampling to produce both faithful and diverse samples. To enhance targeted diversity, we further introduce a Sparse-Aware Semantic Sampling (SASS) strategy to explicitly explore underrepresented regions, improving distribution coverage and sample utility. Extensive experiments on benchmark sEMG datasets, Ninapro DB2, DB4, and DB7, demonstrate that SASG-DA significantly outperforms existing augmentation methods. Overall, our proposed data augmentation approach effectively mitigates overfitting and improves recognition performance and generalization by offering both faithful and diverse samples.
SPRMamba: Surgical Phase Recognition for Endoscopic Submucosal Dissection with Mamba
Sep 18, 2024



Abstract:Endoscopic Submucosal Dissection (ESD) is a minimally invasive procedure initially designed for the treatment of early gastric cancer but is now widely used for various gastrointestinal lesions. Computer-assisted Surgery systems have played a crucial role in improving the precision and safety of ESD procedures, however, their effectiveness is limited by the accurate recognition of surgical phases. The intricate nature of ESD, with different lesion characteristics and tissue structures, presents challenges for real-time surgical phase recognition algorithms. Existing surgical phase recognition algorithms struggle to efficiently capture temporal contexts in video-based scenarios, leading to insufficient performance. To address these issues, we propose SPRMamba, a novel Mamba-based framework for ESD surgical phase recognition. SPRMamba leverages the strengths of Mamba for long-term temporal modeling while introducing the Scaled Residual TranMamba block to enhance the capture of fine-grained details, overcoming the limitations of traditional temporal models like Temporal Convolutional Networks and Transformers. Moreover, a Temporal Sample Strategy is introduced to accelerate the processing, which is essential for real-time phase recognition in clinical settings. Extensive testing on the ESD385 dataset and the cholecystectomy Cholec80 dataset demonstrates that SPRMamba surpasses existing state-of-the-art methods and exhibits greater robustness across various surgical phase recognition tasks.
Deep Multimodal Collaborative Learning for Polyp Re-Identification
Aug 12, 2024Abstract:Colonoscopic Polyp Re-Identification aims to match the same polyp from a large gallery with images from different views taken using different cameras and plays an important role in the prevention and treatment of colorectal cancer in computer-aided diagnosis. However, traditional methods for object ReID directly adopting CNN models trained on the ImageNet dataset usually produce unsatisfactory retrieval performance on colonoscopic datasets due to the large domain gap. Worsely, these solutions typically learn unimodal modal representations on the basis of visual samples, which fails to explore complementary information from different modalities. To address this challenge, we propose a novel Deep Multimodal Collaborative Learning framework named DMCL for polyp re-identification, which can effectively encourage modality collaboration and reinforce generalization capability in medical scenarios. On the basis of it, a dynamic multimodal feature fusion strategy is introduced to leverage the optimized multimodal representations for multimodal fusion via end-to-end training. Experiments on the standard benchmarks show the benefits of the multimodal setting over state-of-the-art unimodal ReID models, especially when combined with the specialized multimodal fusion strategy.
PredIN: Towards Open-Set Gesture Recognition via Prediction Inconsistency
Jul 29, 2024Abstract:Gesture recognition based on surface electromyography (sEMG) has achieved significant progress in human-machine interaction (HMI). However, accurately recognizing predefined gestures within a closed set is still inadequate in practice; a robust open-set system needs to effectively reject unknown gestures while correctly classifying known ones. To handle this challenge, we first report prediction inconsistency discovered for unknown classes due to ensemble diversity, which can significantly facilitate the detection of unknown classes. Based on this insight, we propose an ensemble learning approach, PredIN, to explicitly magnify the prediction inconsistency by enhancing ensemble diversity. Specifically, PredIN maximizes the class feature distribution inconsistency among ensemble members to enhance diversity. Meanwhile, it optimizes inter-class separability within an individual ensemble member to maintain individual performance. Comprehensive experiments on various benchmark datasets demonstrate that the PredIN outperforms state-of-the-art methods by a clear margin.Our proposed method simultaneously achieves accurate closed-set classification for predefined gestures and effective rejection for unknown gestures, exhibiting its efficacy and superiority in open-set gesture recognition based on sEMG.
STS MICCAI 2023 Challenge: Grand challenge on 2D and 3D semi-supervised tooth segmentation
Jul 18, 2024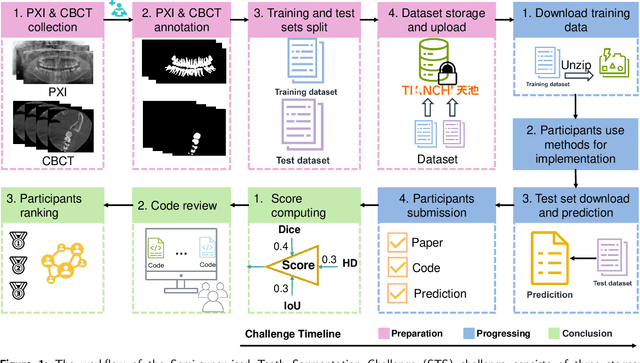
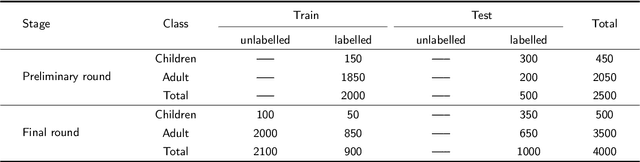
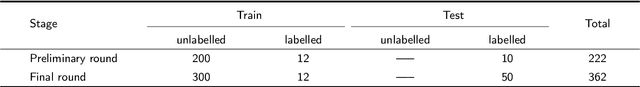
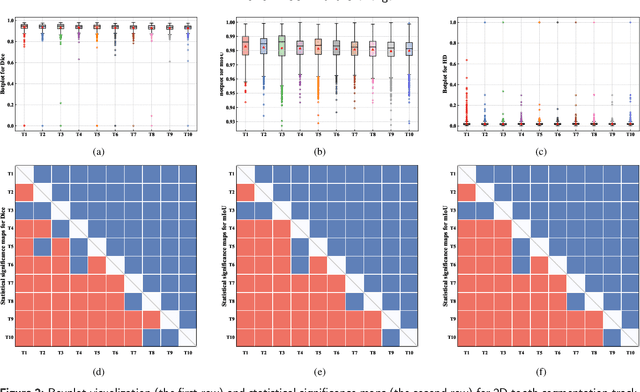
Abstract:Computer-aided design (CAD) tools are increasingly popular in modern dental practice, particularly for treatment planning or comprehensive prognosis evaluation. In particular, the 2D panoramic X-ray image efficiently detects invisible caries, impacted teeth and supernumerary teeth in children, while the 3D dental cone beam computed tomography (CBCT) is widely used in orthodontics and endodontics due to its low radiation dose. However, there is no open-access 2D public dataset for children's teeth and no open 3D dental CBCT dataset, which limits the development of automatic algorithms for segmenting teeth and analyzing diseases. The Semi-supervised Teeth Segmentation (STS) Challenge, a pioneering event in tooth segmentation, was held as a part of the MICCAI 2023 ToothFairy Workshop on the Alibaba Tianchi platform. This challenge aims to investigate effective semi-supervised tooth segmentation algorithms to advance the field of dentistry. In this challenge, we provide two modalities including the 2D panoramic X-ray images and the 3D CBCT tooth volumes. In Task 1, the goal was to segment tooth regions in panoramic X-ray images of both adult and pediatric teeth. Task 2 involved segmenting tooth sections using CBCT volumes. Limited labelled images with mostly unlabelled ones were provided in this challenge prompt using semi-supervised algorithms for training. In the preliminary round, the challenge received registration and result submission by 434 teams, with 64 advancing to the final round. This paper summarizes the diverse methods employed by the top-ranking teams in the STS MICCAI 2023 Challenge.
EDPNet: An Efficient Dual Prototype Network for Motor Imagery EEG Decoding
Jul 03, 2024Abstract:Motor imagery electroencephalograph (MI-EEG) decoding plays a crucial role in developing motor imagery brain-computer interfaces (MI-BCIs). However, decoding intentions from MI remains challenging due to the inherent complexity of EEG signals relative to the small-sample size. In this paper, we propose an Efficient Dual Prototype Network (EDPNet) to enable accurate and fast MI decoding. EDPNet employs a lightweight adaptive spatial-spectral fusion module, which promotes more efficient information fusion between multiple EEG electrodes. Subsequently, a parameter-free multi-scale variance pooling module extracts more comprehensive temporal features. Furthermore, we introduce dual prototypical learning to optimize the feature space distribution and training process, thereby improving the model's generalization ability on small-sample MI datasets. Our experimental results show that the EDPNet outperforms state-of-the-art models with superior classification accuracy and kappa values (84.11% and 0.7881 for dataset BCI competition IV 2a, 86.65% and 0.7330 for dataset BCI competition IV 2b). Additionally, we use the BCI competition III IVa dataset with fewer training data to further validate the generalization ability of the proposed EDPNet. We also achieve superior performance with 82.03% classification accuracy. Benefiting from the lightweight parameters and superior decoding accuracy, our EDPNet shows great potential for MI-BCI applications. The code is publicly available at https://github.com/hancan16/EDPNet.
Hybrid 3D Human Pose Estimation with Monocular Video and Sparse IMUs
Apr 27, 2024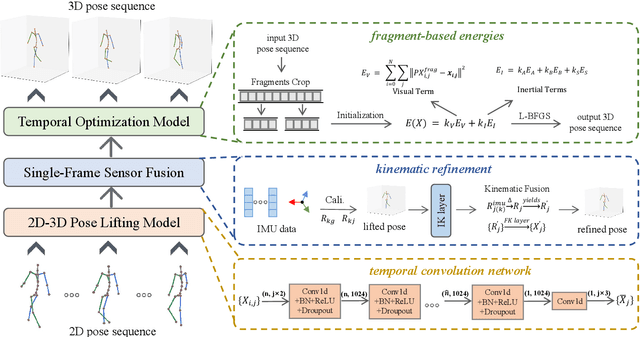
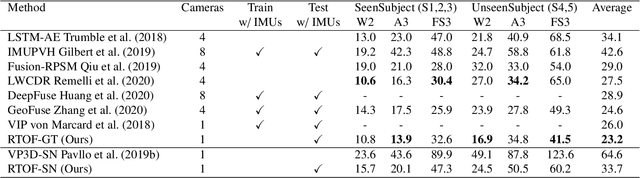
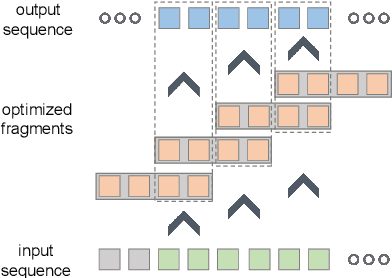
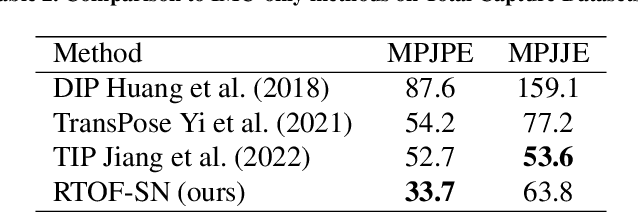
Abstract:Temporal 3D human pose estimation from monocular videos is a challenging task in human-centered computer vision due to the depth ambiguity of 2D-to-3D lifting. To improve accuracy and address occlusion issues, inertial sensor has been introduced to provide complementary source of information. However, it remains challenging to integrate heterogeneous sensor data for producing physically rational 3D human poses. In this paper, we propose a novel framework, Real-time Optimization and Fusion (RTOF), to address this issue. We first incorporate sparse inertial orientations into a parametric human skeleton to refine 3D poses in kinematics. The poses are then optimized by energy functions built on both visual and inertial observations to reduce the temporal jitters. Our framework outputs smooth and biomechanically plausible human motion. Comprehensive experiments with ablation studies demonstrate its rationality and efficiency. On Total Capture dataset, the pose estimation error is significantly decreased compared to the baseline method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge