Chen Liu
Dispersion Loss Counteracts Embedding Condensation and Improves Generalization in Small Language Models
Jan 30, 2026Abstract:Large language models (LLMs) achieve remarkable performance through ever-increasing parameter counts, but scaling incurs steep computational costs. To better understand LLM scaling, we study representational differences between LLMs and their smaller counterparts, with the goal of replicating the representational qualities of larger models in the smaller models. We observe a geometric phenomenon which we term $\textbf{embedding condensation}$, where token embeddings collapse into a narrow cone-like subspace in some language models. Through systematic analyses across multiple Transformer families, we show that small models such as $\texttt{GPT2}$ and $\texttt{Qwen3-0.6B}$ exhibit severe condensation, whereas the larger models such as $\texttt{GPT2-xl}$ and $\texttt{Qwen3-32B}$ are more resistant to this phenomenon. Additional observations show that embedding condensation is not reliably mitigated by knowledge distillation from larger models. To fight against it, we formulate a dispersion loss that explicitly encourages embedding dispersion during training. Experiments demonstrate that it mitigates condensation, recovers dispersion patterns seen in larger models, and yields performance gains across 10 benchmarks. We believe this work offers a principled path toward improving smaller Transformers without additional parameters.
Proof of Time: A Benchmark for Evaluating Scientific Idea Judgments
Jan 12, 2026Abstract:Large language models are increasingly being used to assess and forecast research ideas, yet we lack scalable ways to evaluate the quality of models' judgments about these scientific ideas. Towards this goal, we introduce PoT, a semi-verifiable benchmarking framework that links scientific idea judgments to downstream signals that become observable later (e.g., citations and shifts in researchers' agendas). PoT freezes a pre-cutoff snapshot of evidence in an offline sandbox and asks models to forecast post-cutoff outcomes, enabling verifiable evaluation when ground truth arrives, scalable benchmarking without exhaustive expert annotation, and analysis of human-model misalignment against signals such as peer-review awards. In addition, PoT provides a controlled testbed for agent-based research judgments that evaluate scientific ideas, comparing tool-using agents to non-agent baselines under prompt ablations and budget scaling. Across 30,000+ instances spanning four benchmark domains, we find that, compared with non-agent baselines, higher interaction budgets generally improve agent performance, while the benefit of tool use is strongly task-dependent. By combining time-partitioned, future-verifiable targets with an offline sandbox for tool use, PoT supports scalable evaluation of agents on future-facing scientific idea judgment tasks.
MAI-UI Technical Report: Real-World Centric Foundation GUI Agents
Dec 26, 2025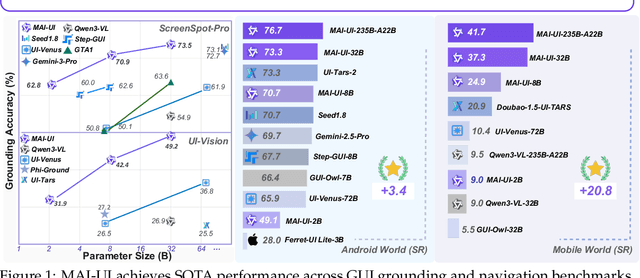

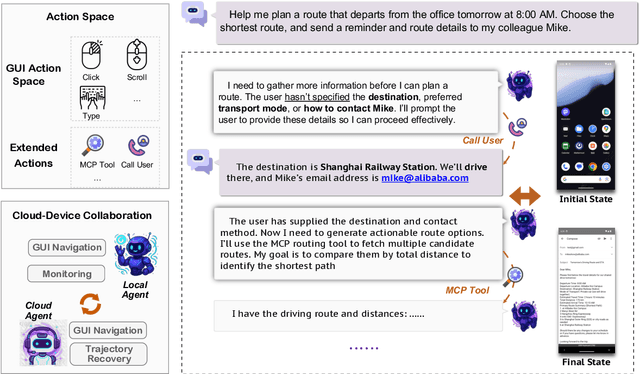
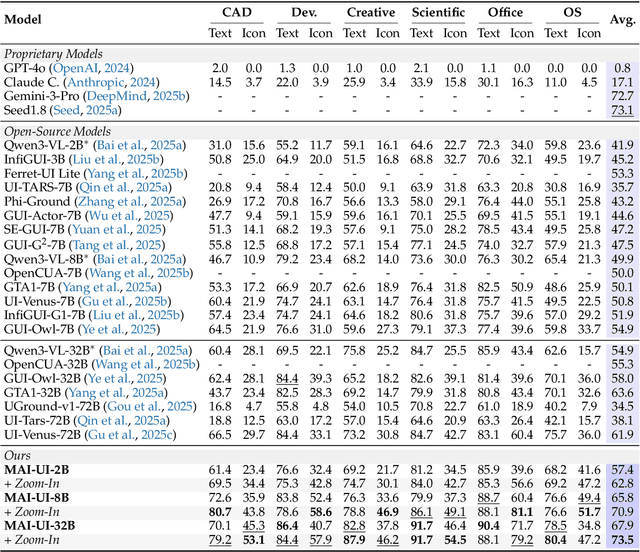
Abstract:The development of GUI agents could revolutionize the next generation of human-computer interaction. Motivated by this vision, we present MAI-UI, a family of foundation GUI agents spanning the full spectrum of sizes, including 2B, 8B, 32B, and 235B-A22B variants. We identify four key challenges to realistic deployment: the lack of native agent-user interaction, the limits of UI-only operation, the absence of a practical deployment architecture, and brittleness in dynamic environments. MAI-UI addresses these issues with a unified methodology: a self-evolving data pipeline that expands the navigation data to include user interaction and MCP tool calls, a native device-cloud collaboration system routes execution by task state, and an online RL framework with advanced optimizations to scale parallel environments and context length. MAI-UI establishes new state-of-the-art across GUI grounding and mobile navigation. On grounding benchmarks, it reaches 73.5% on ScreenSpot-Pro, 91.3% on MMBench GUI L2, 70.9% on OSWorld-G, and 49.2% on UI-Vision, surpassing Gemini-3-Pro and Seed1.8 on ScreenSpot-Pro. On mobile GUI navigation, it sets a new SOTA of 76.7% on AndroidWorld, surpassing UI-Tars-2, Gemini-2.5-Pro and Seed1.8. On MobileWorld, MAI-UI obtains 41.7% success rate, significantly outperforming end-to-end GUI models and competitive with Gemini-3-Pro based agentic frameworks. Our online RL experiments show significant gains from scaling parallel environments from 32 to 512 (+5.2 points) and increasing environment step budget from 15 to 50 (+4.3 points). Finally, the native device-cloud collaboration system improves on-device performance by 33%, reduces cloud model calls by over 40%, and preserves user privacy.
MobileWorld: Benchmarking Autonomous Mobile Agents in Agent-User Interactive, and MCP-Augmented Environments
Dec 22, 2025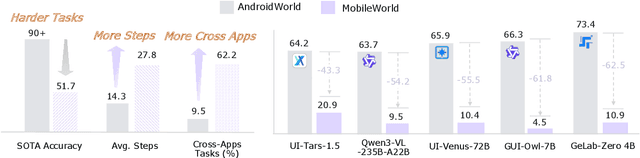
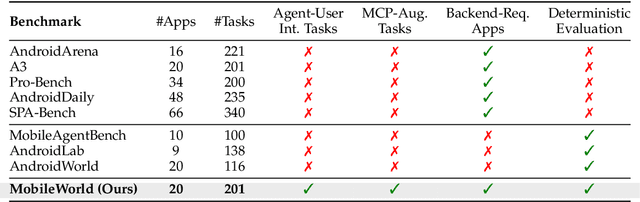
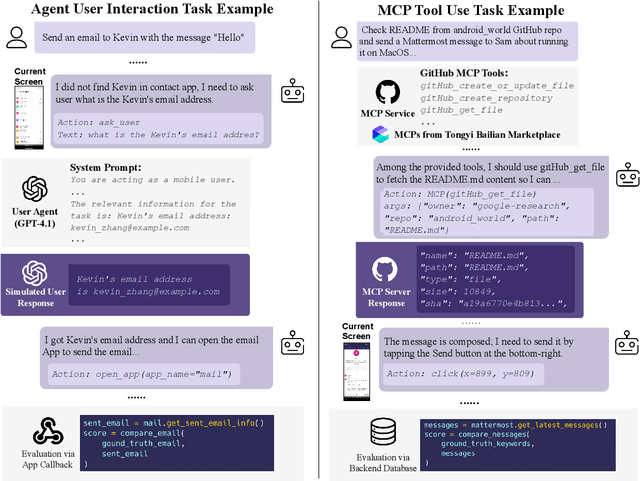

Abstract:Among existing online mobile-use benchmarks, AndroidWorld has emerged as the dominant benchmark due to its reproducible environment and deterministic evaluation; however, recent agents achieving over 90% success rates indicate its saturation and motivate the need for a more challenging benchmark. In addition, its environment lacks key application categories, such as e-commerce and enterprise communication, and does not reflect realistic mobile-use scenarios characterized by vague user instructions and hybrid tool usage. To bridge this gap, we introduce MobileWorld, a substantially more challenging benchmark designed to better reflect real-world mobile usage, comprising 201 tasks across 20 applications, while maintaining the same level of reproducible evaluation as AndroidWorld. The difficulty of MobileWorld is twofold. First, it emphasizes long-horizon tasks with cross-application interactions: MobileWorld requires nearly twice as many task-completion steps on average (27.8 vs. 14.3) and includes far more multi-application tasks (62.2% vs. 9.5%) compared to AndroidWorld. Second, MobileWorld extends beyond standard GUI manipulation by introducing novel task categories, including agent-user interaction and MCP-augmented tasks. To ensure robust evaluation, we provide snapshot-based container environment and precise functional verifications, including backend database inspection and task callback APIs. We further develop a planner-executor agentic framework with extended action spaces to support user interactions and MCP calls. Our results reveal a sharp performance drop compared to AndroidWorld, with the best agentic framework and end-to-end model achieving 51.7% and 20.9% success rates, respectively. Our analysis shows that current models struggle significantly with user interaction and MCP calls, offering a strategic roadmap toward more robust, next-generation mobile intelligence.
Self-Supervised Visual Prompting for Cross-Domain Road Damage Detection
Nov 16, 2025
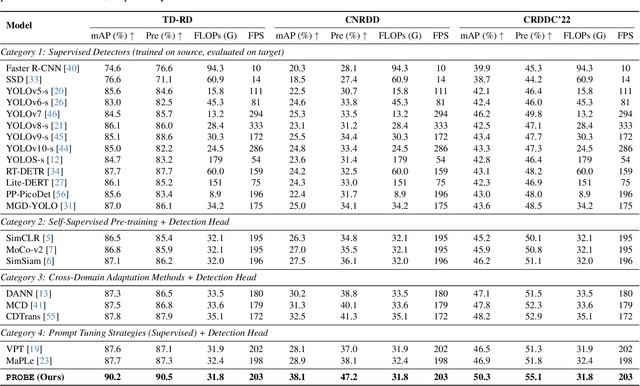
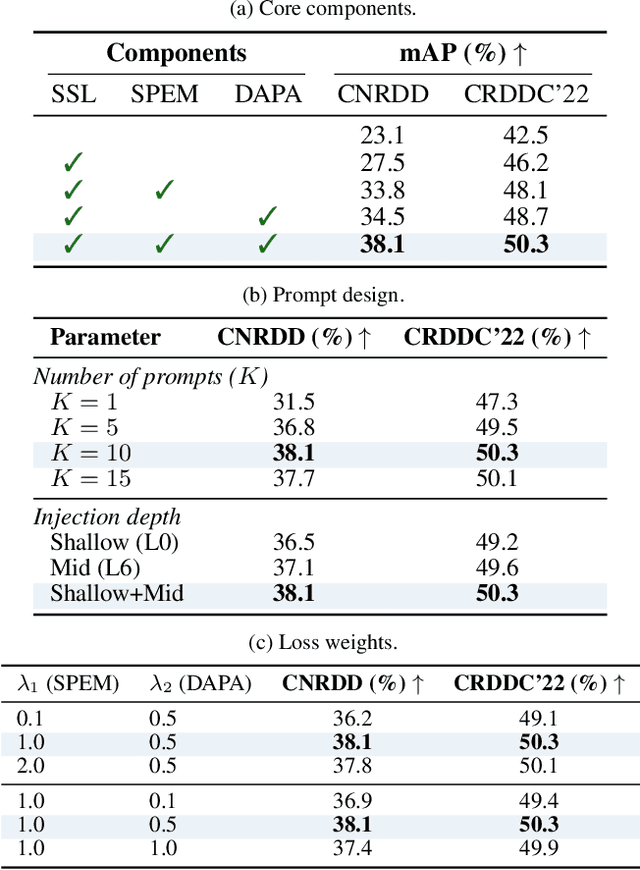

Abstract:The deployment of automated pavement defect detection is often hindered by poor cross-domain generalization. Supervised detectors achieve strong in-domain accuracy but require costly re-annotation for new environments, while standard self-supervised methods capture generic features and remain vulnerable to domain shift. We propose \ours, a self-supervised framework that \emph{visually probes} target domains without labels. \ours introduces a Self-supervised Prompt Enhancement Module (SPEM), which derives defect-aware prompts from unlabeled target data to guide a frozen ViT backbone, and a Domain-Aware Prompt Alignment (DAPA) objective, which aligns prompt-conditioned source and target representations. Experiments on four challenging benchmarks show that \ours consistently outperforms strong supervised, self-supervised, and adaptation baselines, achieving robust zero-shot transfer, improved resilience to domain variations, and high data efficiency in few-shot adaptation. These results highlight self-supervised prompting as a practical direction for building scalable and adaptive visual inspection systems. Source code is publicly available: https://github.com/xixiaouab/PROBE/tree/main
SASG-DA: Sparse-Aware Semantic-Guided Diffusion Augmentation For Myoelectric Gesture Recognition
Nov 12, 2025Abstract:Surface electromyography (sEMG)-based gesture recognition plays a critical role in human-machine interaction (HMI), particularly for rehabilitation and prosthetic control. However, sEMG-based systems often suffer from the scarcity of informative training data, leading to overfitting and poor generalization in deep learning models. Data augmentation offers a promising approach to increasing the size and diversity of training data, where faithfulness and diversity are two critical factors to effectiveness. However, promoting untargeted diversity can result in redundant samples with limited utility. To address these challenges, we propose a novel diffusion-based data augmentation approach, Sparse-Aware Semantic-Guided Diffusion Augmentation (SASG-DA). To enhance generation faithfulness, we introduce the Semantic Representation Guidance (SRG) mechanism by leveraging fine-grained, task-aware semantic representations as generation conditions. To enable flexible and diverse sample generation, we propose a Gaussian Modeling Semantic Sampling (GMSS) strategy, which models the semantic representation distribution and allows stochastic sampling to produce both faithful and diverse samples. To enhance targeted diversity, we further introduce a Sparse-Aware Semantic Sampling (SASS) strategy to explicitly explore underrepresented regions, improving distribution coverage and sample utility. Extensive experiments on benchmark sEMG datasets, Ninapro DB2, DB4, and DB7, demonstrate that SASG-DA significantly outperforms existing augmentation methods. Overall, our proposed data augmentation approach effectively mitigates overfitting and improves recognition performance and generalization by offering both faithful and diverse samples.
Vision Transformer for Robust Occluded Person Reidentification in Complex Surveillance Scenes
Oct 31, 2025Abstract:Person re-identification (ReID) in surveillance is challenged by occlusion, viewpoint distortion, and poor image quality. Most existing methods rely on complex modules or perform well only on clear frontal images. We propose Sh-ViT (Shuffling Vision Transformer), a lightweight and robust model for occluded person ReID. Built on ViT-Base, Sh-ViT introduces three components: First, a Shuffle module in the final Transformer layer to break spatial correlations and enhance robustness to occlusion and blur; Second, scenario-adapted augmentation (geometric transforms, erasing, blur, and color adjustment) to simulate surveillance conditions; Third, DeiT-based knowledge distillation to improve learning with limited labels.To support real-world evaluation, we construct the MyTT dataset, containing over 10,000 pedestrians and 30,000+ images from base station inspections, with frequent equipment occlusion and camera variations. Experiments show that Sh-ViT achieves 83.2% Rank-1 and 80.1% mAP on MyTT, outperforming CNN and ViT baselines, and 94.6% Rank-1 and 87.5% mAP on Market1501, surpassing state-of-the-art methods.In summary, Sh-ViT improves robustness to occlusion and blur without external modules, offering a practical solution for surveillance-based personnel monitoring.
From Prompt Optimization to Multi-Dimensional Credibility Evaluation: Enhancing Trustworthiness of Chinese LLM-Generated Liver MRI Reports
Oct 27, 2025Abstract:Large language models (LLMs) have demonstrated promising performance in generating diagnostic conclusions from imaging findings, thereby supporting radiology reporting, trainee education, and quality control. However, systematic guidance on how to optimize prompt design across different clinical contexts remains underexplored. Moreover, a comprehensive and standardized framework for assessing the trustworthiness of LLM-generated radiology reports is yet to be established. This study aims to enhance the trustworthiness of LLM-generated liver MRI reports by introducing a Multi-Dimensional Credibility Assessment (MDCA) framework and providing guidance on institution-specific prompt optimization. The proposed framework is applied to evaluate and compare the performance of several advanced LLMs, including Kimi-K2-Instruct-0905, Qwen3-235B-A22B-Instruct-2507, DeepSeek-V3, and ByteDance-Seed-OSS-36B-Instruct, using the SiliconFlow platform.
A method for improving multilingual quality and diversity of instruction fine-tuning datasets
Sep 19, 2025Abstract:Multilingual Instruction Fine-Tuning (IFT) is essential for enabling large language models (LLMs) to generalize effectively across diverse linguistic and cultural contexts. However, the scarcity of high-quality multilingual training data and corresponding building method remains a critical bottleneck. While data selection has shown promise in English settings, existing methods often fail to generalize across languages due to reliance on simplistic heuristics or language-specific assumptions. In this work, we introduce Multilingual Data Quality and Diversity (M-DaQ), a novel method for improving LLMs multilinguality, by selecting high-quality and semantically diverse multilingual IFT samples. We further conduct the first systematic investigation of the Superficial Alignment Hypothesis (SAH) in multilingual setting. Empirical results across 18 languages demonstrate that models fine-tuned with M-DaQ method achieve significant performance gains over vanilla baselines over 60% win rate. Human evaluations further validate these gains, highlighting the increment of cultural points in the response. We release the M-DaQ code to support future research.
Gradient Inversion Transcript: Leveraging Robust Generative Priors to Reconstruct Training Data from Gradient Leakage
May 26, 2025Abstract:We propose Gradient Inversion Transcript (GIT), a novel generative approach for reconstructing training data from leaked gradients. GIT employs a generative attack model, whose architecture is tailored to align with the structure of the leaked model based on theoretical analysis. Once trained offline, GIT can be deployed efficiently and only relies on the leaked gradients to reconstruct the input data, rendering it applicable under various distributed learning environments. When used as a prior for other iterative optimization-based methods, GIT not only accelerates convergence but also enhances the overall reconstruction quality. GIT consistently outperforms existing methods across multiple datasets and demonstrates strong robustness under challenging conditions, including inaccurate gradients, data distribution shifts and discrepancies in model parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge