Liang Hong
Rank-and-Reason: Multi-Agent Collaboration Accelerates Zero-Shot Protein Mutation Prediction
Feb 03, 2026Abstract:Zero-shot mutation prediction is vital for low-resource protein engineering, yet existing protein language models (PLMs) often yield statistically confident results that ignore fundamental biophysical constraints. Currently, selecting candidates for wet-lab validation relies on manual expert auditing of PLM outputs, a process that is inefficient, subjective, and highly dependent on domain expertise. To address this, we propose Rank-and-Reason (VenusRAR), a two-stage agentic framework to automate this workflow and maximize expected wet-lab fitness. In the Rank-Stage, a Computational Expert and Virtual Biologist aggregate a context-aware multi-modal ensemble, establishing a new Spearman correlation record of 0.551 (vs. 0.518) on ProteinGym. In the Reason-Stage, an agentic Expert Panel employs chain-of-thought reasoning to audit candidates against geometric and structural constraints, improving the Top-5 Hit Rate by up to 367% on ProteinGym-DMS99. The wet-lab validation on Cas12i3 nuclease further confirms the framework's efficacy, achieving a 46.7% positive rate and identifying two novel mutants with 4.23-fold and 5.05-fold activity improvements. Code and datasets are released on GitHub (https://github.com/ai4protein/VenusRAR/).
A new strategy for finite-sample valid prediction of future insurance claims in the regression setting
Jan 29, 2026Abstract:The extant insurance literature demonstrates a paucity of finite-sample valid prediction intervals of future insurance claims in the regression setting. To address this challenge, this article proposes a new strategy that converts a predictive method in the unsupervised iid (independent identically distributed) setting to a predictive method in the regression setting. In particular, it enables an actuary to obtain infinitely many finite-sample valid prediction intervals in the regression setting.
VenusX: Unlocking Fine-Grained Functional Understanding of Proteins
May 17, 2025



Abstract:Deep learning models have driven significant progress in predicting protein function and interactions at the protein level. While these advancements have been invaluable for many biological applications such as enzyme engineering and function annotation, a more detailed perspective is essential for understanding protein functional mechanisms and evaluating the biological knowledge captured by models. To address this demand, we introduce VenusX, the first large-scale benchmark for fine-grained functional annotation and function-based protein pairing at the residue, fragment, and domain levels. VenusX comprises three major task categories across six types of annotations, including residue-level binary classification, fragment-level multi-class classification, and pairwise functional similarity scoring for identifying critical active sites, binding sites, conserved sites, motifs, domains, and epitopes. The benchmark features over 878,000 samples curated from major open-source databases such as InterPro, BioLiP, and SAbDab. By providing mixed-family and cross-family splits at three sequence identity thresholds, our benchmark enables a comprehensive assessment of model performance on both in-distribution and out-of-distribution scenarios. For baseline evaluation, we assess a diverse set of popular and open-source models, including pre-trained protein language models, sequence-structure hybrids, structure-based methods, and alignment-based techniques. Their performance is reported across all benchmark datasets and evaluation settings using multiple metrics, offering a thorough comparison and a strong foundation for future research. Code and data are publicly available at https://github.com/ai4protein/VenusX.
VenusFactory: A Unified Platform for Protein Engineering Data Retrieval and Language Model Fine-Tuning
Mar 19, 2025Abstract:Natural language processing (NLP) has significantly influenced scientific domains beyond human language, including protein engineering, where pre-trained protein language models (PLMs) have demonstrated remarkable success. However, interdisciplinary adoption remains limited due to challenges in data collection, task benchmarking, and application. This work presents VenusFactory, a versatile engine that integrates biological data retrieval, standardized task benchmarking, and modular fine-tuning of PLMs. VenusFactory supports both computer science and biology communities with choices of both a command-line execution and a Gradio-based no-code interface, integrating $40+$ protein-related datasets and $40+$ popular PLMs. All implementations are open-sourced on https://github.com/tyang816/VenusFactory.
Finite-sample valid prediction of future insurance claims in the regression problem
Mar 05, 2025Abstract:In the current insurance literature, prediction of insurance claims in the regression problem is often performed with a statistical model. This model-based approach may suffer from several drawbacks: (i) model misspecification, (ii) selection effect, and (iii) lack of finite-sample validity. This article addresses these three issues simultaneously by employing conformal prediction-a general machine learning strategy for valid predictions. The proposed method is both model-free and tuning-parameter-free. It also guarantees finite-sample validity at a pre-assigned coverage probability level.
AI-Driven Secure Data Sharing: A Trustworthy and Privacy-Preserving Approach
Jan 26, 2025



Abstract:In the era of data-driven decision-making, ensuring the privacy and security of shared data is paramount across various domains. Applying existing deep neural networks (DNNs) to encrypted data is critical and often compromises performance, security, and computational overhead. To address these limitations, this research introduces a secure framework consisting of a learnable encryption method based on the block-pixel operation to encrypt the data and subsequently integrate it with the Vision Transformer (ViT). The proposed framework ensures data privacy and security by creating unique scrambling patterns per key, providing robust performance against adversarial attacks without compromising computational efficiency and data integrity. The framework was tested on sensitive medical datasets to validate its efficacy, proving its ability to handle highly confidential information securely. The suggested framework was validated with a 94\% success rate after extensive testing on real-world datasets, such as MRI brain tumors and histological scans of lung and colon cancers. Additionally, the framework was tested under diverse adversarial attempts against secure data sharing with optimum performance and demonstrated its effectiveness in various threat scenarios. These comprehensive analyses underscore its robustness, making it a trustworthy solution for secure data sharing in critical applications.
Retrieval-Enhanced Mutation Mastery: Augmenting Zero-Shot Prediction of Protein Language Model
Oct 28, 2024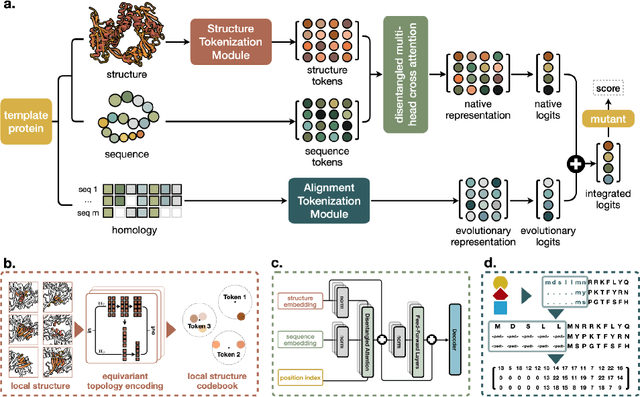
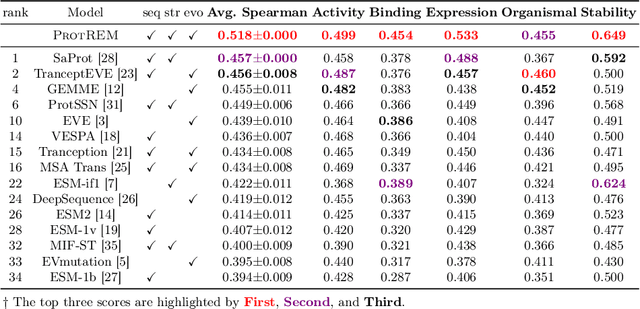
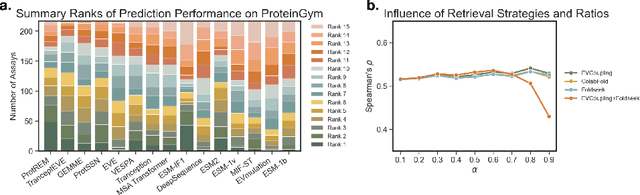
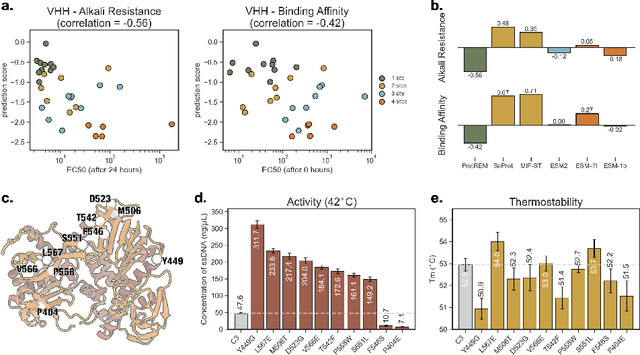
Abstract:Enzyme engineering enables the modification of wild-type proteins to meet industrial and research demands by enhancing catalytic activity, stability, binding affinities, and other properties. The emergence of deep learning methods for protein modeling has demonstrated superior results at lower costs compared to traditional approaches such as directed evolution and rational design. In mutation effect prediction, the key to pre-training deep learning models lies in accurately interpreting the complex relationships among protein sequence, structure, and function. This study introduces a retrieval-enhanced protein language model for comprehensive analysis of native properties from sequence and local structural interactions, as well as evolutionary properties from retrieved homologous sequences. The state-of-the-art performance of the proposed ProtREM is validated on over 2 million mutants across 217 assays from an open benchmark (ProteinGym). We also conducted post-hoc analyses of the model's ability to improve the stability and binding affinity of a VHH antibody. Additionally, we designed 10 new mutants on a DNA polymerase and conducted wet-lab experiments to evaluate their enhanced activity at higher temperatures. Both in silico and experimental evaluations confirmed that our method provides reliable predictions of mutation effects, offering an auxiliary tool for biologists aiming to evolve existing enzymes. The implementation is publicly available at https://github.com/tyang816/ProtREM.
Advancing Healthcare: Innovative ML Approaches for Improved Medical Imaging in Data-Constrained Environments
Oct 16, 2024



Abstract:Healthcare industries face challenges when experiencing rare diseases due to limited samples. Artificial Intelligence (AI) communities overcome this situation to create synthetic data which is an ethical and privacy issue in the medical domain. This research introduces the CAT-U-Net framework as a new approach to overcome these limitations, which enhances feature extraction from medical images without the need for large datasets. The proposed framework adds an extra concatenation layer with downsampling parts, thereby improving its ability to learn from limited data while maintaining patient privacy. To validate, the proposed framework's robustness, different medical conditioning datasets were utilized including COVID-19, brain tumors, and wrist fractures. The framework achieved nearly 98% reconstruction accuracy, with a Dice coefficient close to 0.946. The proposed CAT-U-Net has the potential to make a big difference in medical image diagnostics in settings with limited data.
Immunogenicity Prediction with Dual Attention Enables Vaccine Target Selection
Oct 03, 2024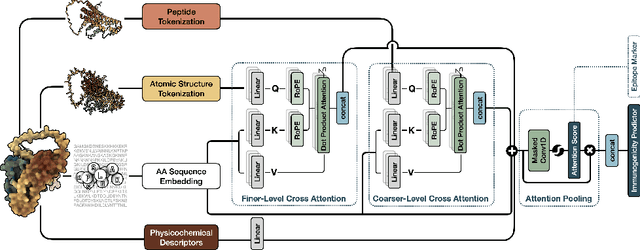
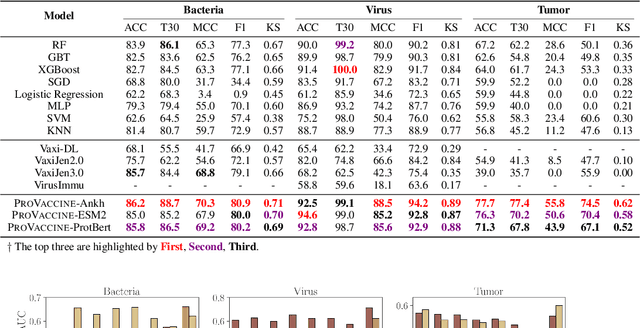
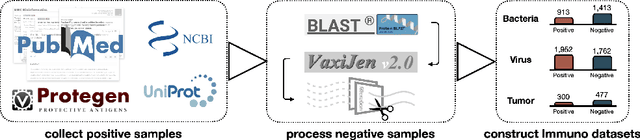

Abstract:Immunogenicity prediction is a central topic in reverse vaccinology for finding candidate vaccines that can trigger protective immune responses. Existing approaches typically rely on highly compressed features and simple model architectures, leading to limited prediction accuracy and poor generalizability. To address these challenges, we introduce ProVaccine, a novel deep learning solution with a dual attention mechanism that integrates pre-trained latent vector representations of protein sequences and structures. We also compile the most comprehensive immunogenicity dataset to date, encompassing over 9,500 antigen sequences, structures, and immunogenicity labels from bacteria, viruses, and tumors. Extensive experiments demonstrate that ProVaccine outperforms existing methods across a wide range of evaluation metrics. Furthermore, we establish a post-hoc validation protocol to assess the practical significance of deep learning models in tackling vaccine design challenges. Our work provides an effective tool for vaccine design and sets valuable benchmarks for future research.
Reactzyme: A Benchmark for Enzyme-Reaction Prediction
Aug 24, 2024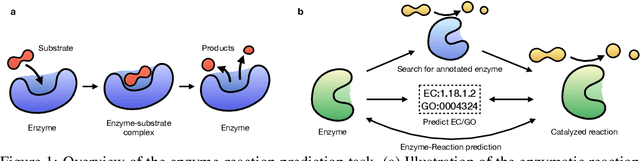
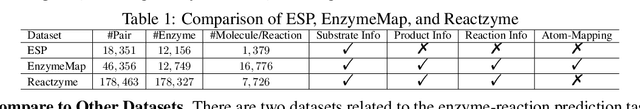
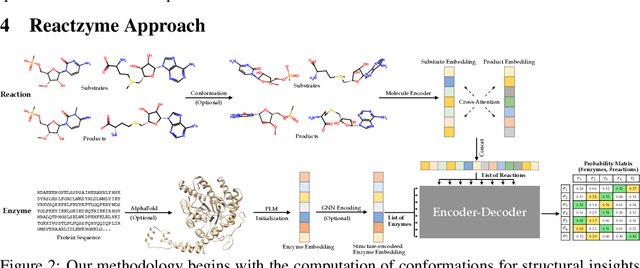
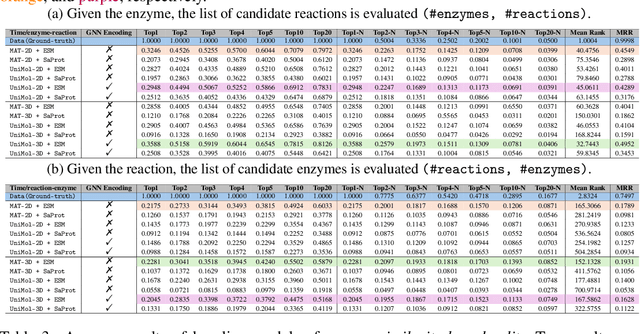
Abstract:Enzymes, with their specific catalyzed reactions, are necessary for all aspects of life, enabling diverse biological processes and adaptations. Predicting enzyme functions is essential for understanding biological pathways, guiding drug development, enhancing bioproduct yields, and facilitating evolutionary studies. Addressing the inherent complexities, we introduce a new approach to annotating enzymes based on their catalyzed reactions. This method provides detailed insights into specific reactions and is adaptable to newly discovered reactions, diverging from traditional classifications by protein family or expert-derived reaction classes. We employ machine learning algorithms to analyze enzyme reaction datasets, delivering a much more refined view on the functionality of enzymes. Our evaluation leverages the largest enzyme-reaction dataset to date, derived from the SwissProt and Rhea databases with entries up to January 8, 2024. We frame the enzyme-reaction prediction as a retrieval problem, aiming to rank enzymes by their catalytic ability for specific reactions. With our model, we can recruit proteins for novel reactions and predict reactions in novel proteins, facilitating enzyme discovery and function annotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge