Lirong Zheng
Fourier-RWKV: A Multi-State Perception Network for Efficient Image Dehazing
Dec 09, 2025Abstract:Image dehazing is crucial for reliable visual perception, yet it remains highly challenging under real-world non-uniform haze conditions. Although Transformer-based methods excel at capturing global context, their quadratic computational complexity hinders real-time deployment. To address this, we propose Fourier Receptance Weighted Key Value (Fourier-RWKV), a novel dehazing framework based on a Multi-State Perception paradigm. The model achieves comprehensive haze degradation modeling with linear complexity by synergistically integrating three distinct perceptual states: (1) Spatial-form Perception, realized through the Deformable Quad-directional Token Shift (DQ-Shift) operation, which dynamically adjusts receptive fields to accommodate local haze variations; (2) Frequency-domain Perception, implemented within the Fourier Mix block, which extends the core WKV attention mechanism of RWKV from the spatial domain to the Fourier domain, preserving the long-range dependencies essential for global haze estimation while mitigating spatial attenuation; (3) Semantic-relation Perception, facilitated by the Semantic Bridge Module (SBM), which utilizes Dynamic Semantic Kernel Fusion (DSK-Fusion) to precisely align encoder-decoder features and suppress artifacts. Extensive experiments on multiple benchmarks demonstrate that Fourier-RWKV delivers state-of-the-art performance across diverse haze scenarios while significantly reducing computational overhead, establishing a favorable trade-off between restoration quality and practical efficiency. Code is available at: https://github.com/Dilizlr/Fourier-RWKV.
Secondary Structure-Guided Novel Protein Sequence Generation with Latent Graph Diffusion
Jul 10, 2024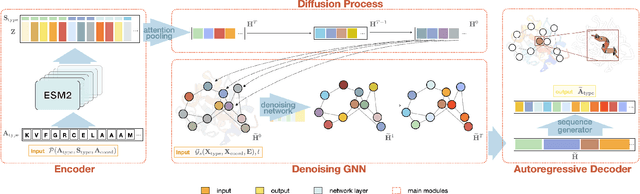
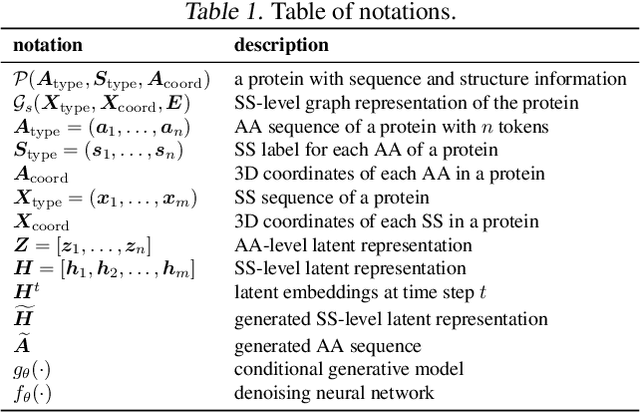
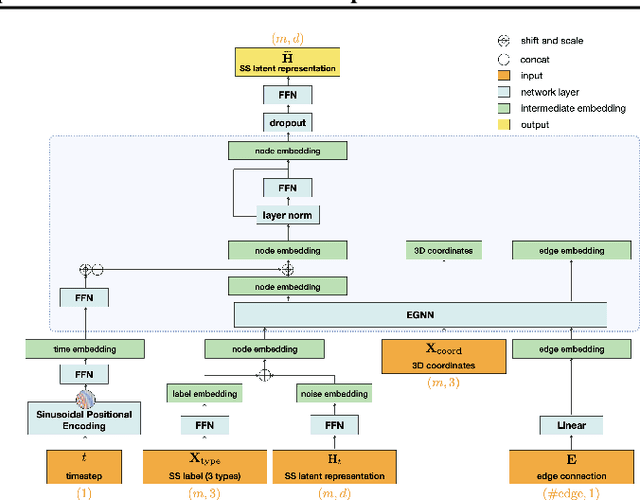

Abstract:The advent of deep learning has introduced efficient approaches for de novo protein sequence design, significantly improving success rates and reducing development costs compared to computational or experimental methods. However, existing methods face challenges in generating proteins with diverse lengths and shapes while maintaining key structural features. To address these challenges, we introduce CPDiffusion-SS, a latent graph diffusion model that generates protein sequences based on coarse-grained secondary structural information. CPDiffusion-SS offers greater flexibility in producing a variety of novel amino acid sequences while preserving overall structural constraints, thus enhancing the reliability and diversity of generated proteins. Experimental analyses demonstrate the significant superiority of the proposed method in producing diverse and novel sequences, with CPDiffusion-SS surpassing popular baseline methods on open benchmarks across various quantitative measurements. Furthermore, we provide a series of case studies to highlight the biological significance of the generation performance by the proposed method. The source code is publicly available at https://github.com/riacd/CPDiffusion-SS
Protein Representation Learning with Sequence Information Embedding: Does it Always Lead to a Better Performance?
Jun 28, 2024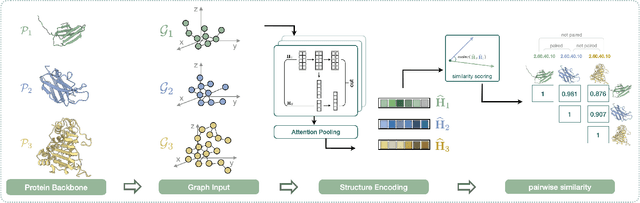
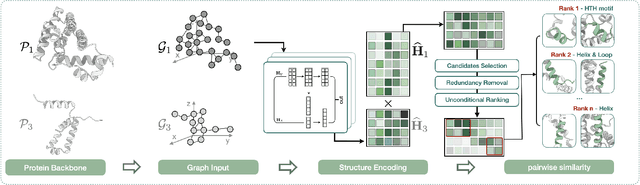

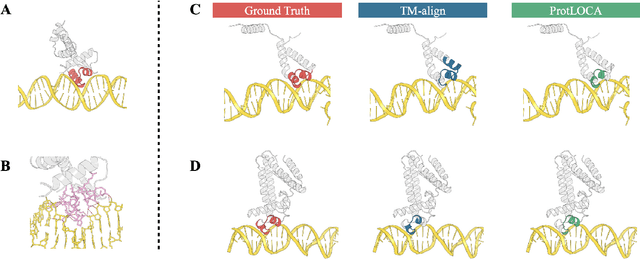
Abstract:Deep learning has become a crucial tool in studying proteins. While the significance of modeling protein structure has been discussed extensively in the literature, amino acid types are typically included in the input as a default operation for many inference tasks. This study demonstrates with structure alignment task that embedding amino acid types in some cases may not help a deep learning model learn better representation. To this end, we propose ProtLOCA, a local geometry alignment method based solely on amino acid structure representation. The effectiveness of ProtLOCA is examined by a global structure-matching task on protein pairs with an independent test dataset based on CATH labels. Our method outperforms existing sequence- and structure-based representation learning methods by more quickly and accurately matching structurally consistent protein domains. Furthermore, in local structure pairing tasks, ProtLOCA for the first time provides a valid solution to highlight common local structures among proteins with different overall structures but the same function. This suggests a new possibility for using deep learning methods to analyze protein structure to infer function.
Simple, Efficient and Scalable Structure-aware Adapter Boosts Protein Language Models
Apr 23, 2024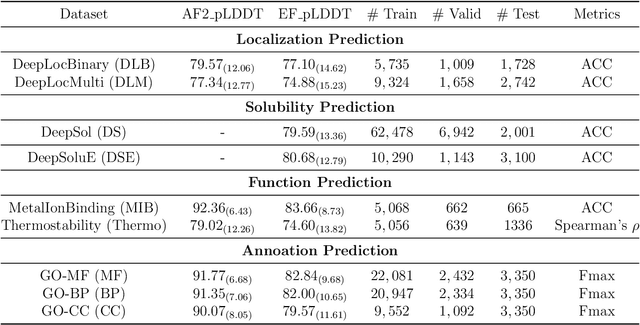
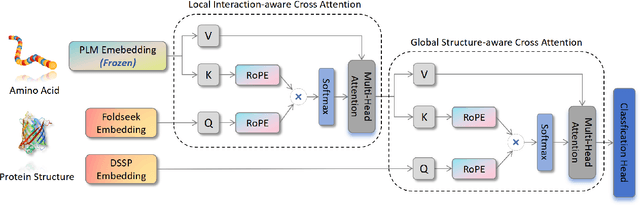
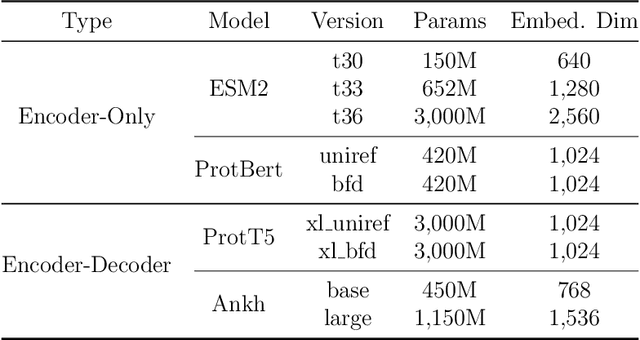
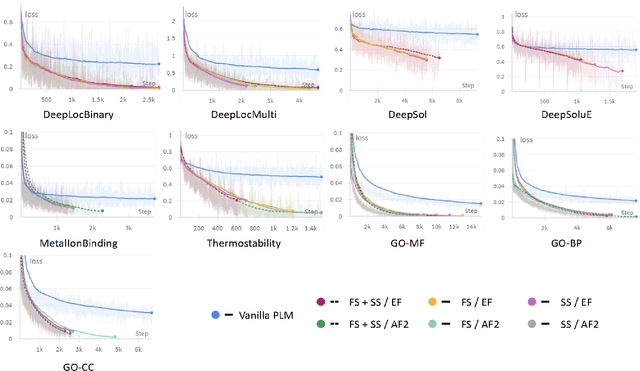
Abstract:Fine-tuning Pre-trained protein language models (PLMs) has emerged as a prominent strategy for enhancing downstream prediction tasks, often outperforming traditional supervised learning approaches. As a widely applied powerful technique in natural language processing, employing Parameter-Efficient Fine-Tuning techniques could potentially enhance the performance of PLMs. However, the direct transfer to life science tasks is non-trivial due to the different training strategies and data forms. To address this gap, we introduce SES-Adapter, a simple, efficient, and scalable adapter method for enhancing the representation learning of PLMs. SES-Adapter incorporates PLM embeddings with structural sequence embeddings to create structure-aware representations. We show that the proposed method is compatible with different PLM architectures and across diverse tasks. Extensive evaluations are conducted on 2 types of folding structures with notable quality differences, 9 state-of-the-art baselines, and 9 benchmark datasets across distinct downstream tasks. Results show that compared to vanilla PLMs, SES-Adapter improves downstream task performance by a maximum of 11% and an average of 3%, with significantly accelerated training speed by a maximum of 1034% and an average of 362%, the convergence rate is also improved by approximately 2 times. Moreover, positive optimization is observed even with low-quality predicted structures. The source code for SES-Adapter is available at https://github.com/tyang816/SES-Adapter.
A Unified View on Neural Message Passing with Opinion Dynamics for Social Networks
Oct 03, 2023Abstract:Social networks represent a common form of interconnected data frequently depicted as graphs within the domain of deep learning-based inference. These communities inherently form dynamic systems, achieving stability through continuous internal communications and opinion exchanges among social actors along their social ties. In contrast, neural message passing in deep learning provides a clear and intuitive mathematical framework for understanding information propagation and aggregation among connected nodes in graphs. Node representations are dynamically updated by considering both the connectivity and status of neighboring nodes. This research harmonizes concepts from sociometry and neural message passing to analyze and infer the behavior of dynamic systems. Drawing inspiration from opinion dynamics in sociology, we propose ODNet, a novel message passing scheme incorporating bounded confidence, to refine the influence weight of local nodes for message propagation. We adjust the similarity cutoffs of bounded confidence and influence weights of ODNet and define opinion exchange rules that align with the characteristics of social network graphs. We show that ODNet enhances prediction performance across various graph types and alleviates oversmoothing issues. Furthermore, our approach surpasses conventional baselines in graph representation learning and proves its practical significance in analyzing real-world co-occurrence networks of metabolic genes. Remarkably, our method simplifies complex social network graphs solely by leveraging knowledge of interaction frequencies among entities within the system. It accurately identifies internal communities and the roles of genes in different metabolic pathways, including opinion leaders, bridge communicators, and isolators.
Factor Decomposed Generative Adversarial Networks for Text-to-Image Synthesis
Mar 24, 2023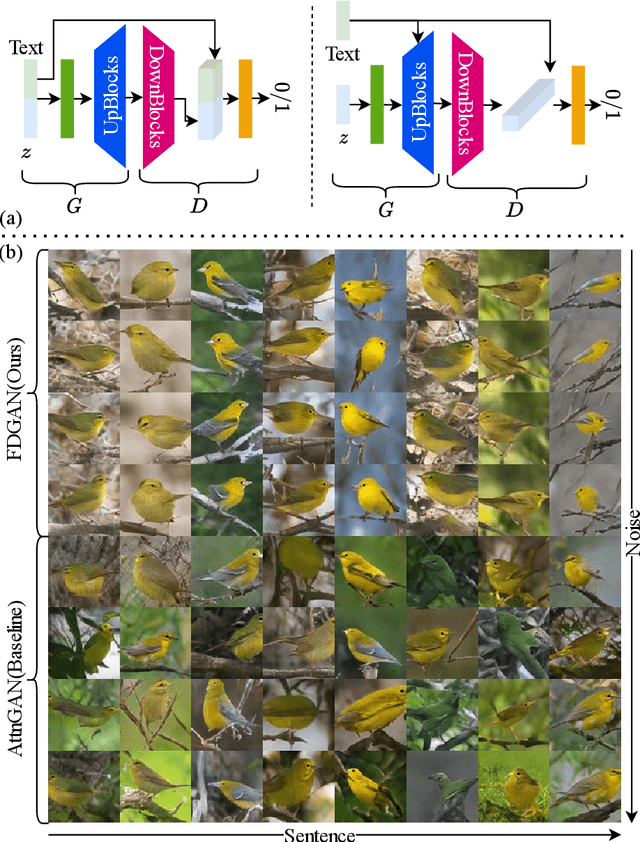

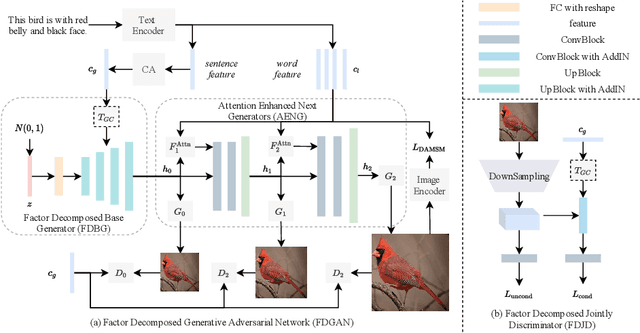

Abstract:Prior works about text-to-image synthesis typically concatenated the sentence embedding with the noise vector, while the sentence embedding and the noise vector are two different factors, which control the different aspects of the generation. Simply concatenating them will entangle the latent factors and encumber the generative model. In this paper, we attempt to decompose these two factors and propose Factor Decomposed Generative Adversarial Networks~(FDGAN). To achieve this, we firstly generate images from the noise vector and then apply the sentence embedding in the normalization layer for both generator and discriminators. We also design an additive norm layer to align and fuse the text-image features. The experimental results show that decomposing the noise and the sentence embedding can disentangle latent factors in text-to-image synthesis, and make the generative model more efficient. Compared with the baseline, FDGAN can achieve better performance, while fewer parameters are used.
Time-frequency Network for Robust Speaker Recognition
Mar 07, 2023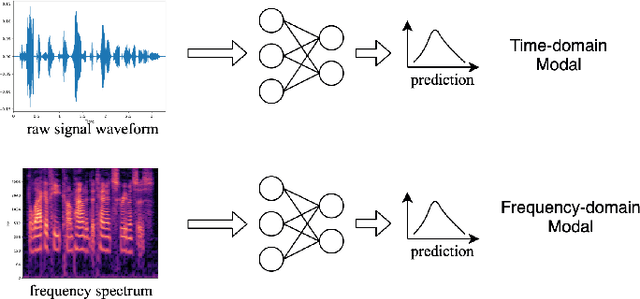
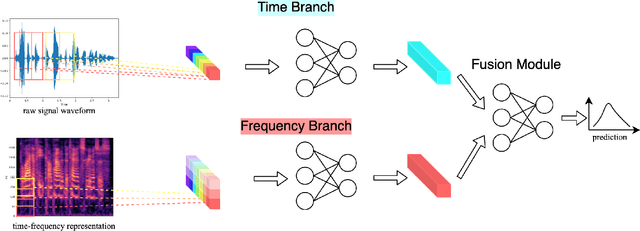
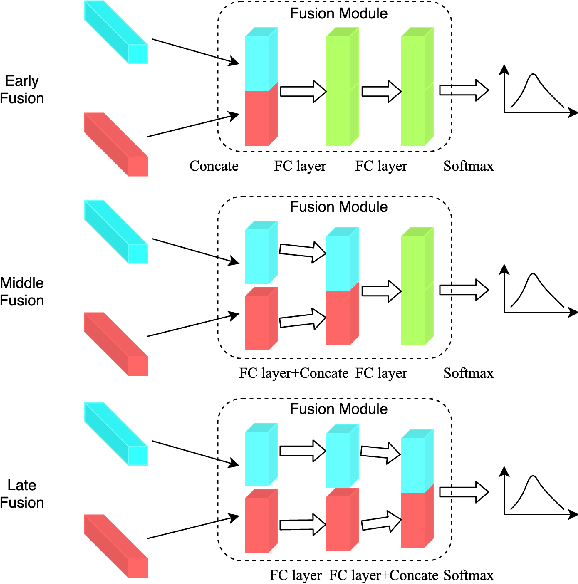
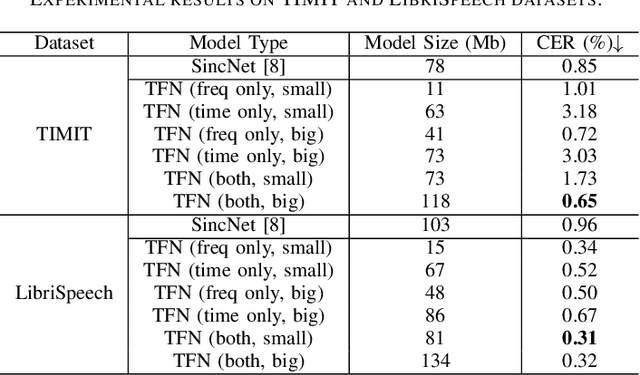
Abstract:The wide deployment of speech-based biometric systems usually demands high-performance speaker recognition algorithms. However, most of the prior works for speaker recognition either process the speech in the frequency domain or time domain, which may produce suboptimal results because both time and frequency domains are important for speaker recognition. In this paper, we attempt to analyze the speech signal in both time and frequency domains and propose the time-frequency network~(TFN) for speaker recognition by extracting and fusing the features in the two domains. Based on the recent advance of deep neural networks, we propose a convolution neural network to encode the raw speech waveform and the frequency spectrum into domain-specific features, which are then fused and transformed into a classification feature space for speaker recognition. Experimental results on the publicly available datasets TIMIT and LibriSpeech show that our framework is effective to combine the information in the two domains and performs better than the state-of-the-art methods for speaker recognition.
A Ligand-and-structure Dual-driven Deep Learning Method for the Discovery of Highly Potent GnRH1R Antagonist to treat Uterine Diseases
Jul 23, 2022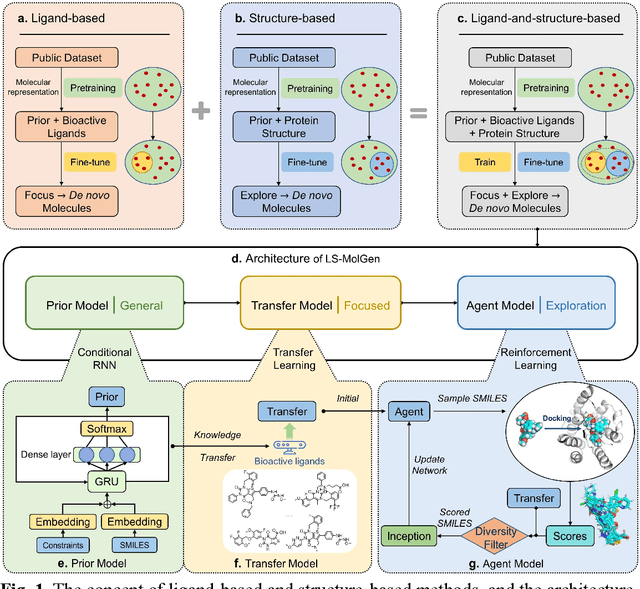
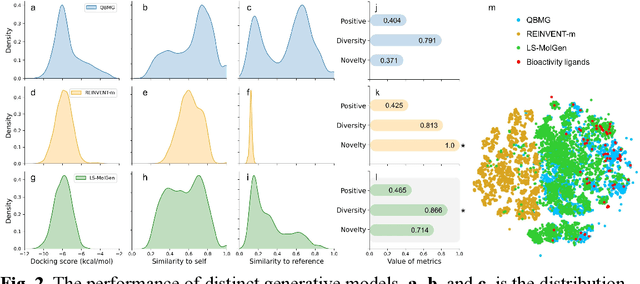
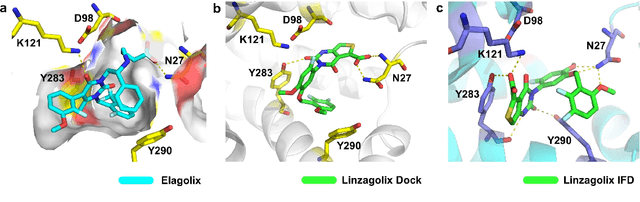
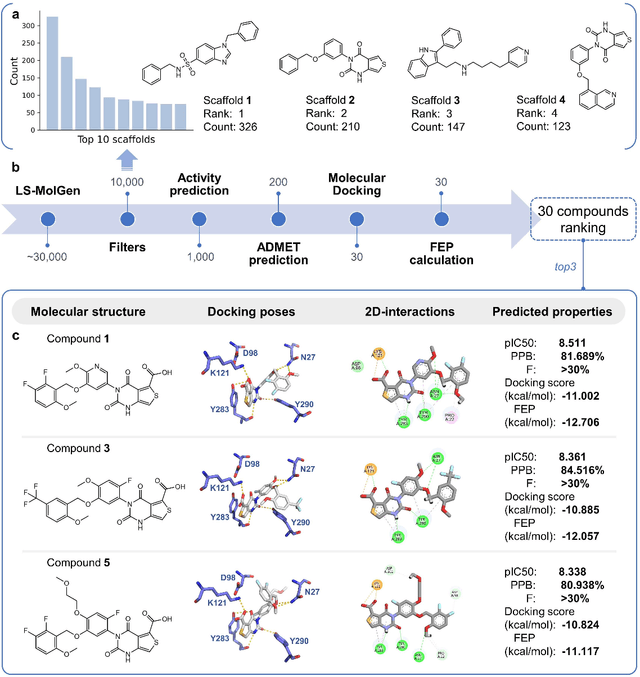
Abstract:Gonadotrophin-releasing hormone receptor (GnRH1R) is a promising therapeutic target for the treatment of uterine diseases. To date, several GnRH1R antagonists are available in clinical investigation without satisfying multiple property constraints. To fill this gap, we aim to develop a deep learning-based framework to facilitate the effective and efficient discovery of a new orally active small-molecule drug targeting GnRH1R with desirable properties. In the present work, a ligand-and-structure combined model, namely LS-MolGen, was firstly proposed for molecular generation by fully utilizing the information on the known active compounds and the structure of the target protein, which was demonstrated by its superior performance than ligand- or structure-based methods separately. Then, a in silico screening including activity prediction, ADMET evaluation, molecular docking and FEP calculation was conducted, where ~30,000 generated novel molecules were narrowed down to 8 for experimental synthesis and validation. In vitro and in vivo experiments showed that three of them exhibited potent inhibition activities (compound 5 IC50 = 0.856 nM, compound 6 IC50 = 0.901 nM, compound 7 IC50 = 2.54 nM) against GnRH1R, and compound 5 performed well in fundamental PK properties, such as half-life, oral bioavailability, and PPB, etc. We believed that the proposed ligand-and-structure combined molecular generative model and the whole computer-aided workflow can potentially be extended to similar tasks for de novo drug design or lead optimization.
T-Net: Deep Stacked Scale-Iteration Network for Image Dehazing
Jun 05, 2021
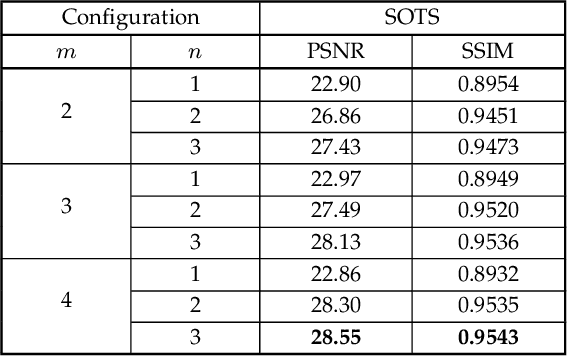
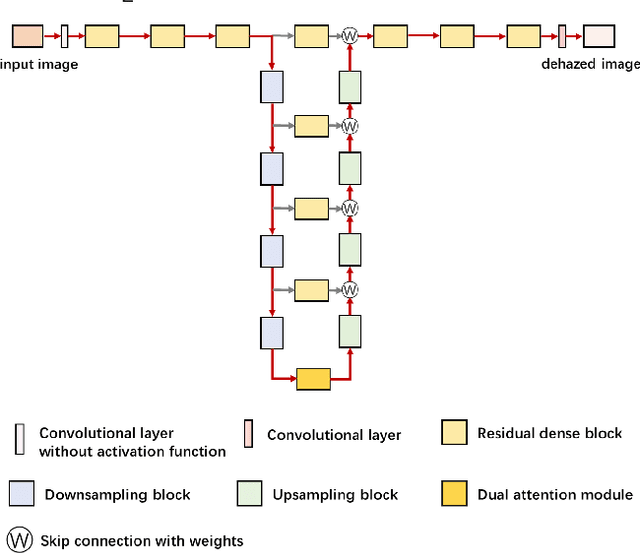
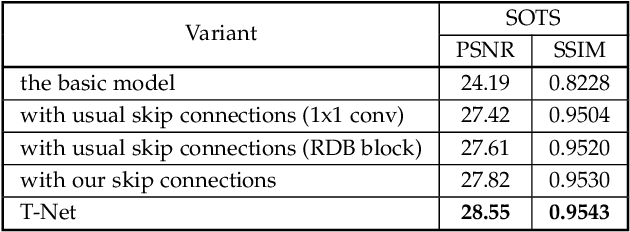
Abstract:Hazy images reduce the visibility of the image content, and haze will lead to failure in handling subsequent computer vision tasks. In this paper, we address the problem of image dehazing by proposing a dehazing network named T-Net, which consists of a backbone network based on the U-Net architecture and a dual attention module. And it can achieve multi-scale feature fusion by using skip connections with a new fusion strategy. Furthermore, by repeatedly unfolding the plain T-Net, Stack T-Net is proposed to take advantage of the dependence of deep features across stages via a recursive strategy. In order to reduce network parameters, the intra-stage recursive computation of ResNet is adopted in our Stack T-Net. And we take both the stage-wise result and the original hazy image as input to each T-Net and finally output the prediction of clean image. Experimental results on both synthetic and real-world images demonstrate that our plain T-Net and the advanced Stack T-Net perform favorably against the state-of-the-art dehazing algorithms, and show that our Stack T-Net could further improve the dehazing effect, demonstrating the effectiveness of the recursive strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge