Mingchen Li
Rank-and-Reason: Multi-Agent Collaboration Accelerates Zero-Shot Protein Mutation Prediction
Feb 03, 2026Abstract:Zero-shot mutation prediction is vital for low-resource protein engineering, yet existing protein language models (PLMs) often yield statistically confident results that ignore fundamental biophysical constraints. Currently, selecting candidates for wet-lab validation relies on manual expert auditing of PLM outputs, a process that is inefficient, subjective, and highly dependent on domain expertise. To address this, we propose Rank-and-Reason (VenusRAR), a two-stage agentic framework to automate this workflow and maximize expected wet-lab fitness. In the Rank-Stage, a Computational Expert and Virtual Biologist aggregate a context-aware multi-modal ensemble, establishing a new Spearman correlation record of 0.551 (vs. 0.518) on ProteinGym. In the Reason-Stage, an agentic Expert Panel employs chain-of-thought reasoning to audit candidates against geometric and structural constraints, improving the Top-5 Hit Rate by up to 367% on ProteinGym-DMS99. The wet-lab validation on Cas12i3 nuclease further confirms the framework's efficacy, achieving a 46.7% positive rate and identifying two novel mutants with 4.23-fold and 5.05-fold activity improvements. Code and datasets are released on GitHub (https://github.com/ai4protein/VenusRAR/).
GLEN-Bench: A Graph-Language based Benchmark for Nutritional Health
Jan 26, 2026Abstract:Nutritional interventions are important for managing chronic health conditions, but current computational methods provide limited support for personalized dietary guidance. We identify three key gaps: (1) dietary pattern studies often ignore real-world constraints such as socioeconomic status, comorbidities, and limited food access; (2) recommendation systems rarely explain why a particular food helps a given patient; and (3) no unified benchmark evaluates methods across the connected tasks needed for nutritional interventions. We introduce GLEN-Bench, the first comprehensive graph-language based benchmark for nutritional health assessment. We combine NHANES health records, FNDDS food composition data, and USDA food-access metrics to build a knowledge graph that links demographics, health conditions, dietary behaviors, poverty-related constraints, and nutrient needs. We test the benchmark using opioid use disorder, where models must detect subtle nutritional differences across disease stages. GLEN-Bench includes three linked tasks: risk detection identifies at-risk individuals from dietary and socioeconomic patterns; recommendation suggests personalized foods that meet clinical needs within resource constraints; and question answering provides graph-grounded, natural-language explanations to facilitate comprehension. We evaluate these graph-language approaches, including graph neural networks, large language models, and hybrid architectures, to establish solid baselines and identify practical design choices. Our analysis identifies clear dietary patterns linked to health risks, providing insights that can guide practical interventions.
NL2Repo-Bench: Towards Long-Horizon Repository Generation Evaluation of Coding Agents
Dec 14, 2025Abstract:Recent advances in coding agents suggest rapid progress toward autonomous software development, yet existing benchmarks fail to rigorously evaluate the long-horizon capabilities required to build complete software systems. Most prior evaluations focus on localized code generation, scaffolded completion, or short-term repair tasks, leaving open the question of whether agents can sustain coherent reasoning, planning, and execution over the extended horizons demanded by real-world repository construction. To address this gap, we present NL2Repo Bench, a benchmark explicitly designed to evaluate the long-horizon repository generation ability of coding agents. Given only a single natural-language requirements document and an empty workspace, agents must autonomously design the architecture, manage dependencies, implement multi-module logic, and produce a fully installable Python library. Our experiments across state-of-the-art open- and closed-source models reveal that long-horizon repository generation remains largely unsolved: even the strongest agents achieve below 40% average test pass rates and rarely complete an entire repository correctly. Detailed analysis uncovers fundamental long-horizon failure modes, including premature termination, loss of global coherence, fragile cross-file dependencies, and inadequate planning over hundreds of interaction steps. NL2Repo Bench establishes a rigorous, verifiable testbed for measuring sustained agentic competence and highlights long-horizon reasoning as a central bottleneck for the next generation of autonomous coding agents.
Hollywood Town: Long-Video Generation via Cross-Modal Multi-Agent Orchestration
Oct 25, 2025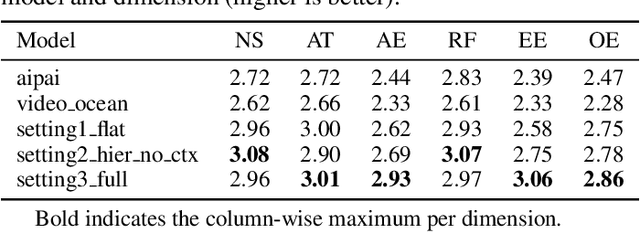
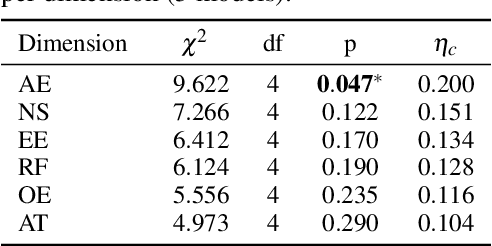
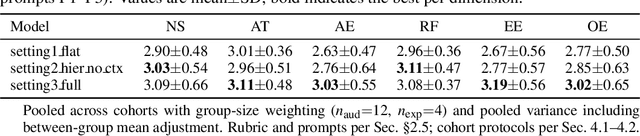
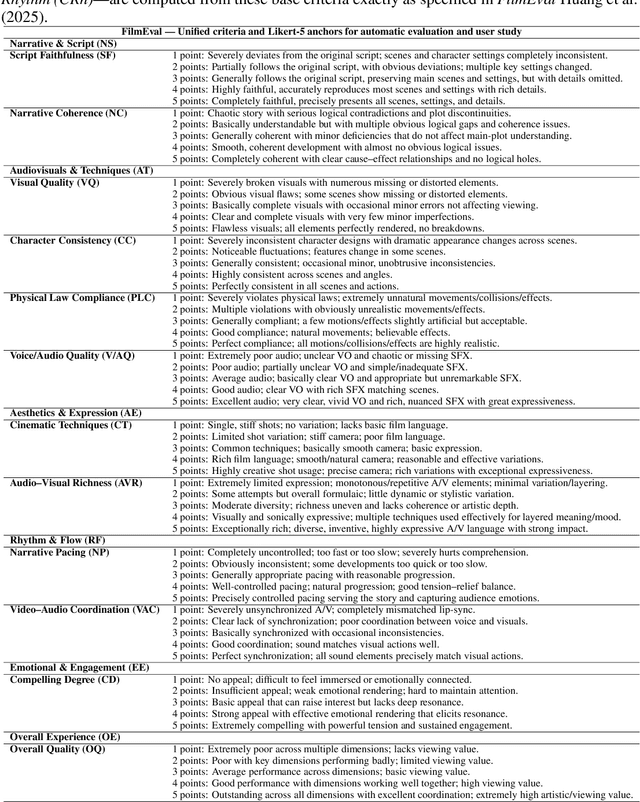
Abstract:Recent advancements in multi-agent systems have demonstrated significant potential for enhancing creative task performance, such as long video generation. This study introduces three innovations to improve multi-agent collaboration. First, we propose OmniAgent, a hierarchical, graph-based multi-agent framework for long video generation that leverages a film-production-inspired architecture to enable modular specialization and scalable inter-agent collaboration. Second, inspired by context engineering, we propose hypergraph nodes that enable temporary group discussions among agents lacking sufficient context, reducing individual memory requirements while ensuring adequate contextual information. Third, we transition from directed acyclic graphs (DAGs) to directed cyclic graphs with limited retries, allowing agents to reflect and refine outputs iteratively, thereby improving earlier stages through feedback from subsequent nodes. These contributions lay the groundwork for developing more robust multi-agent systems in creative tasks.
JALMBench: Benchmarking Jailbreak Vulnerabilities in Audio Language Models
May 23, 2025


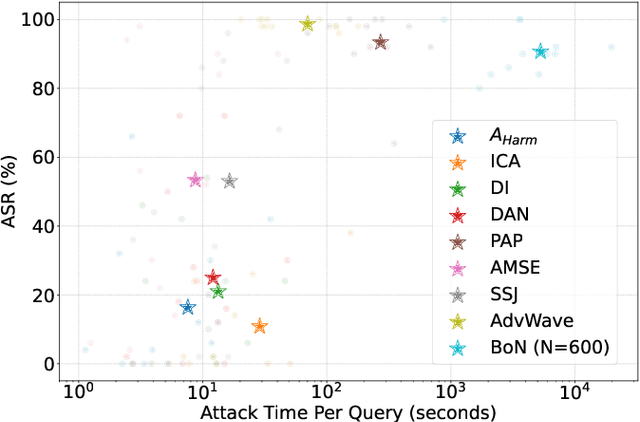
Abstract:Audio Language Models (ALMs) have made significant progress recently. These models integrate the audio modality directly into the model, rather than converting speech into text and inputting text to Large Language Models (LLMs). While jailbreak attacks on LLMs have been extensively studied, the security of ALMs with audio modalities remains largely unexplored. Currently, there is a lack of an adversarial audio dataset and a unified framework specifically designed to evaluate and compare attacks and ALMs. In this paper, we present JALMBench, the \textit{first} comprehensive benchmark to assess the safety of ALMs against jailbreak attacks. JALMBench includes a dataset containing 2,200 text samples and 51,381 audio samples with over 268 hours. It supports 12 mainstream ALMs, 4 text-transferred and 4 audio-originated attack methods, and 5 defense methods. Using JALMBench, we provide an in-depth analysis of attack efficiency, topic sensitivity, voice diversity, and attack representations. Additionally, we explore mitigation strategies for the attacks at both the prompt level and the response level.
Exploiting Meta-Learning-based Poisoning Attacks for Graph Link Prediction
Apr 08, 2025



Abstract:Link prediction in graph data utilizes various algorithms and machine learning/deep learning models to predict potential relationships between graph nodes. This technique has found widespread use in numerous real-world applications, including recommendation systems, community networks, and biological structures. However, recent research has highlighted the vulnerability of link prediction models to adversarial attacks, such as poisoning and evasion attacks. Addressing the vulnerability of these models is crucial to ensure stable and robust performance in link prediction applications. While many works have focused on enhancing the robustness of the Graph Convolution Network (GCN) model, the Variational Graph Auto-Encoder (VGAE), a sophisticated model for link prediction, has not been thoroughly investigated in the context of graph adversarial attacks. To bridge this gap, this article proposes an unweighted graph poisoning attack approach using meta-learning techniques to undermine VGAE's link prediction performance. We conducted comprehensive experiments on diverse datasets to evaluate the proposed method and its parameters, comparing it with existing approaches in similar settings. Our results demonstrate that our approach significantly diminishes link prediction performance and outperforms other state-of-the-art methods.
VenusFactory: A Unified Platform for Protein Engineering Data Retrieval and Language Model Fine-Tuning
Mar 19, 2025Abstract:Natural language processing (NLP) has significantly influenced scientific domains beyond human language, including protein engineering, where pre-trained protein language models (PLMs) have demonstrated remarkable success. However, interdisciplinary adoption remains limited due to challenges in data collection, task benchmarking, and application. This work presents VenusFactory, a versatile engine that integrates biological data retrieval, standardized task benchmarking, and modular fine-tuning of PLMs. VenusFactory supports both computer science and biology communities with choices of both a command-line execution and a Gradio-based no-code interface, integrating $40+$ protein-related datasets and $40+$ popular PLMs. All implementations are open-sourced on https://github.com/tyang816/VenusFactory.
TVNet: A Novel Time Series Analysis Method Based on Dynamic Convolution and 3D-Variation
Mar 10, 2025



Abstract:With the recent development and advancement of Transformer and MLP architectures, significant strides have been made in time series analysis. Conversely, the performance of Convolutional Neural Networks (CNNs) in time series analysis has fallen short of expectations, diminishing their potential for future applications. Our research aims to enhance the representational capacity of Convolutional Neural Networks (CNNs) in time series analysis by introducing novel perspectives and design innovations. To be specific, We introduce a novel time series reshaping technique that considers the inter-patch, intra-patch, and cross-variable dimensions. Consequently, we propose TVNet, a dynamic convolutional network leveraging a 3D perspective to employ time series analysis. TVNet retains the computational efficiency of CNNs and achieves state-of-the-art results in five key time series analysis tasks, offering a superior balance of efficiency and performance over the state-of-the-art Transformer-based and MLP-based models. Additionally, our findings suggest that TVNet exhibits enhanced transferability and robustness. Therefore, it provides a new perspective for applying CNN in advanced time series analysis tasks.
DP-GTR: Differentially Private Prompt Protection via Group Text Rewriting
Mar 06, 2025



Abstract:Prompt privacy is crucial, especially when using online large language models (LLMs), due to the sensitive information often contained within prompts. While LLMs can enhance prompt privacy through text rewriting, existing methods primarily focus on document-level rewriting, neglecting the rich, multi-granular representations of text. This limitation restricts LLM utilization to specific tasks, overlooking their generalization and in-context learning capabilities, thus hindering practical application. To address this gap, we introduce DP-GTR, a novel three-stage framework that leverages local differential privacy (DP) and the composition theorem via group text rewriting. DP-GTR is the first framework to integrate both document-level and word-level information while exploiting in-context learning to simultaneously improve privacy and utility, effectively bridging local and global DP mechanisms at the individual data point level. Experiments on CommonSense QA and DocVQA demonstrate that DP-GTR outperforms existing approaches, achieving a superior privacy-utility trade-off. Furthermore, our framework is compatible with existing rewriting techniques, serving as a plug-in to enhance privacy protection. Our code is publicly available at https://github.com/FatShion-FTD/DP-GTR for reproducibility.
Automatic Pruning via Structured Lasso with Class-wise Information
Feb 13, 2025Abstract:Most pruning methods concentrate on unimportant filters of neural networks. However, they face the loss of statistical information due to a lack of consideration for class-wise data. In this paper, from the perspective of leveraging precise class-wise information for model pruning, we utilize structured lasso with guidance from Information Bottleneck theory. Our approach ensures that statistical information is retained during the pruning process. With these techniques, we introduce two innovative adaptive network pruning schemes: sparse graph-structured lasso pruning with Information Bottleneck (\textbf{sGLP-IB}) and sparse tree-guided lasso pruning with Information Bottleneck (\textbf{sTLP-IB}). The key aspect is pruning model filters using sGLP-IB and sTLP-IB to better capture class-wise relatedness. Compared to multiple state-of-the-art methods, our approaches demonstrate superior performance across three datasets and six model architectures in extensive experiments. For instance, using the VGG16 model on the CIFAR-10 dataset, we achieve a parameter reduction of 85%, a decrease in FLOPs by 61%, and maintain an accuracy of 94.10% (0.14% higher than the original model); we reduce the parameters by 55% with the accuracy at 76.12% using the ResNet architecture on ImageNet (only drops 0.03%). In summary, we successfully reduce model size and computational resource usage while maintaining accuracy. Our codes are at https://anonymous.4open.science/r/IJCAI-8104.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge