Zaifu Zhan
Data-Efficient Biomedical In-Context Learning: A Diversity-Enhanced Submodular Perspective
Aug 11, 2025Abstract:Recent progress in large language models (LLMs) has leveraged their in-context learning (ICL) abilities to enable quick adaptation to unseen biomedical NLP tasks. By incorporating only a few input-output examples into prompts, LLMs can rapidly perform these new tasks. While the impact of these demonstrations on LLM performance has been extensively studied, most existing approaches prioritize representativeness over diversity when selecting examples from large corpora. To address this gap, we propose Dual-Div, a diversity-enhanced data-efficient framework for demonstration selection in biomedical ICL. Dual-Div employs a two-stage retrieval and ranking process: First, it identifies a limited set of candidate examples from a corpus by optimizing both representativeness and diversity (with optional annotation for unlabeled data). Second, it ranks these candidates against test queries to select the most relevant and non-redundant demonstrations. Evaluated on three biomedical NLP tasks (named entity recognition (NER), relation extraction (RE), and text classification (TC)) using LLaMA 3.1 and Qwen 2.5 for inference, along with three retrievers (BGE-Large, BMRetriever, MedCPT), Dual-Div consistently outperforms baselines-achieving up to 5% higher macro-F1 scores-while demonstrating robustness to prompt permutations and class imbalance. Our findings establish that diversity in initial retrieval is more critical than ranking-stage optimization, and limiting demonstrations to 3-5 examples maximizes performance efficiency.
Automating Expert-Level Medical Reasoning Evaluation of Large Language Models
Jul 10, 2025Abstract:As large language models (LLMs) become increasingly integrated into clinical decision-making, ensuring transparent and trustworthy reasoning is essential. However, existing evaluation strategies of LLMs' medical reasoning capability either suffer from unsatisfactory assessment or poor scalability, and a rigorous benchmark remains lacking. To address this, we introduce MedThink-Bench, a benchmark designed for rigorous, explainable, and scalable assessment of LLMs' medical reasoning. MedThink-Bench comprises 500 challenging questions across ten medical domains, each annotated with expert-crafted step-by-step rationales. Building on this, we propose LLM-w-Ref, a novel evaluation framework that leverages fine-grained rationales and LLM-as-a-Judge mechanisms to assess intermediate reasoning with expert-level fidelity while maintaining scalability. Experiments show that LLM-w-Ref exhibits a strong positive correlation with expert judgments. Benchmarking twelve state-of-the-art LLMs, we find that smaller models (e.g., MedGemma-27B) can surpass larger proprietary counterparts (e.g., OpenAI-o3). Overall, MedThink-Bench offers a foundational tool for evaluating LLMs' medical reasoning, advancing their safe and responsible deployment in clinical practice.
Uncertainty-Aware Large Language Models for Explainable Disease Diagnosis
May 06, 2025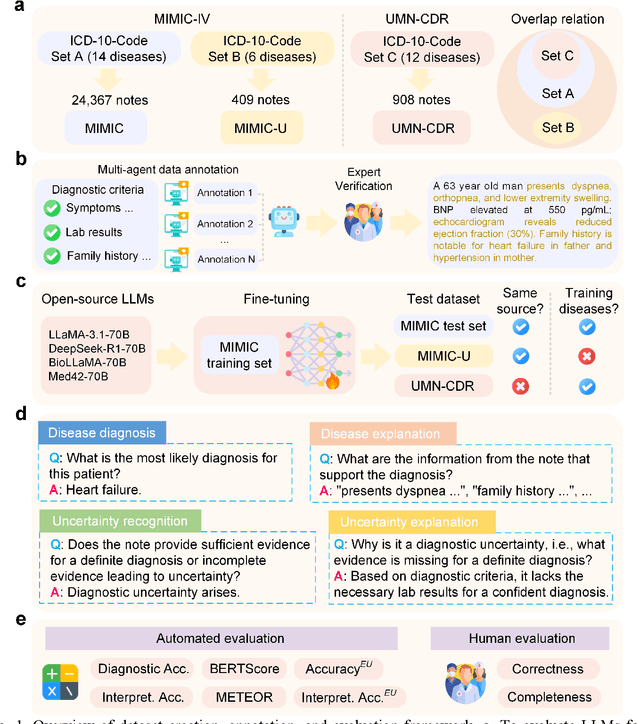
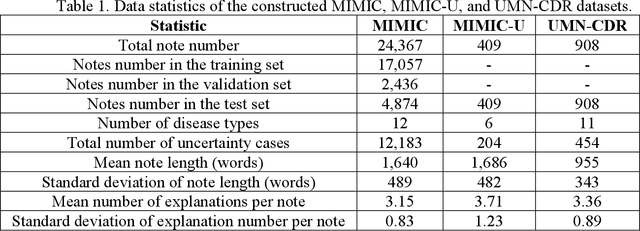
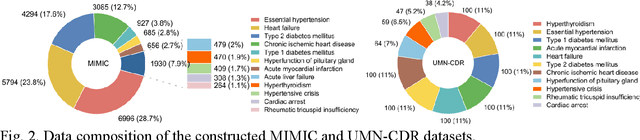
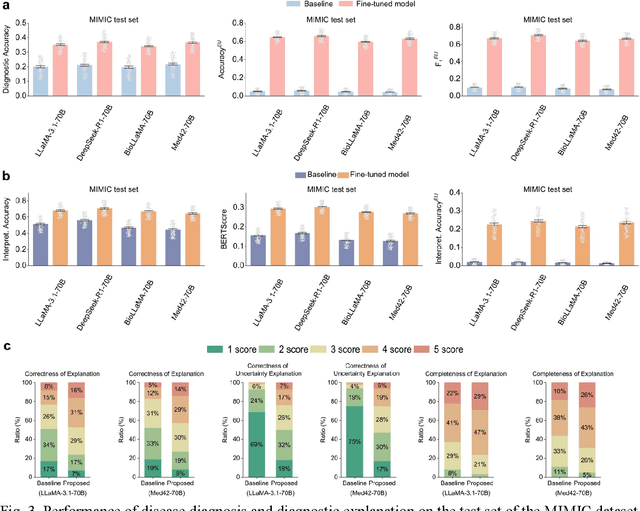
Abstract:Explainable disease diagnosis, which leverages patient information (e.g., signs and symptoms) and computational models to generate probable diagnoses and reasonings, offers clear clinical values. However, when clinical notes encompass insufficient evidence for a definite diagnosis, such as the absence of definitive symptoms, diagnostic uncertainty usually arises, increasing the risk of misdiagnosis and adverse outcomes. Although explicitly identifying and explaining diagnostic uncertainties is essential for trustworthy diagnostic systems, it remains under-explored. To fill this gap, we introduce ConfiDx, an uncertainty-aware large language model (LLM) created by fine-tuning open-source LLMs with diagnostic criteria. We formalized the task and assembled richly annotated datasets that capture varying degrees of diagnostic ambiguity. Evaluating ConfiDx on real-world datasets demonstrated that it excelled in identifying diagnostic uncertainties, achieving superior diagnostic performance, and generating trustworthy explanations for diagnoses and uncertainties. To our knowledge, this is the first study to jointly address diagnostic uncertainty recognition and explanation, substantially enhancing the reliability of automatic diagnostic systems.
Retrieval-augmented in-context learning for multimodal large language models in disease classification
May 04, 2025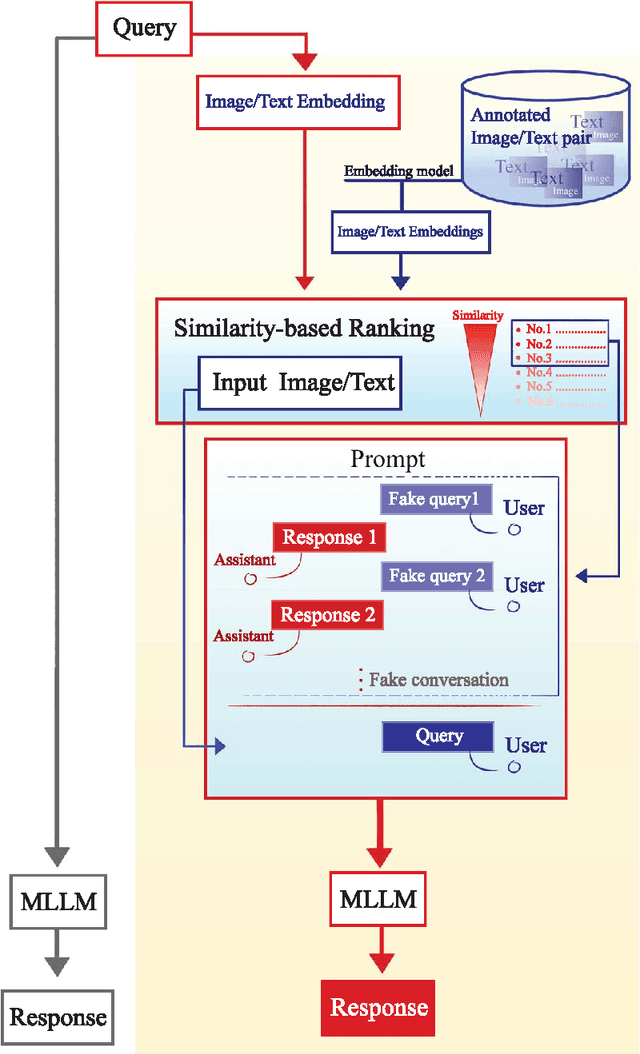
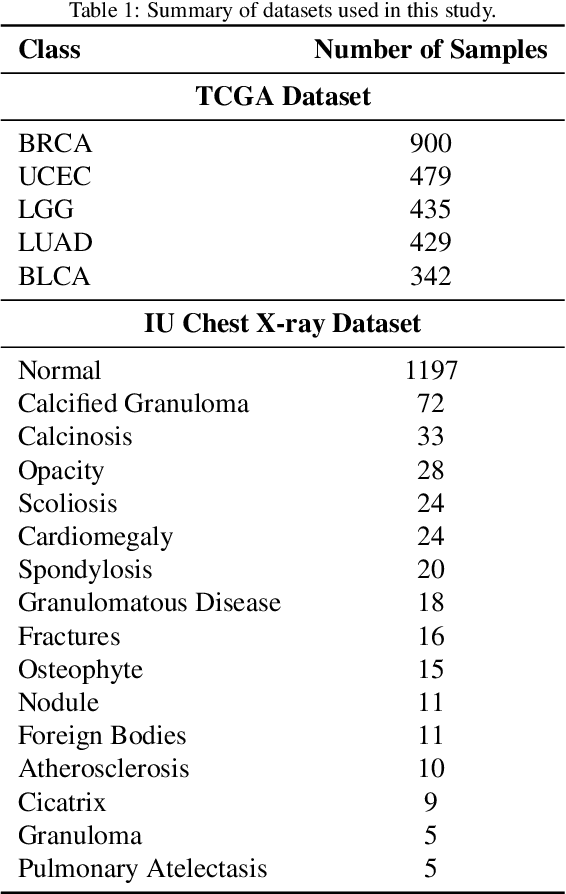
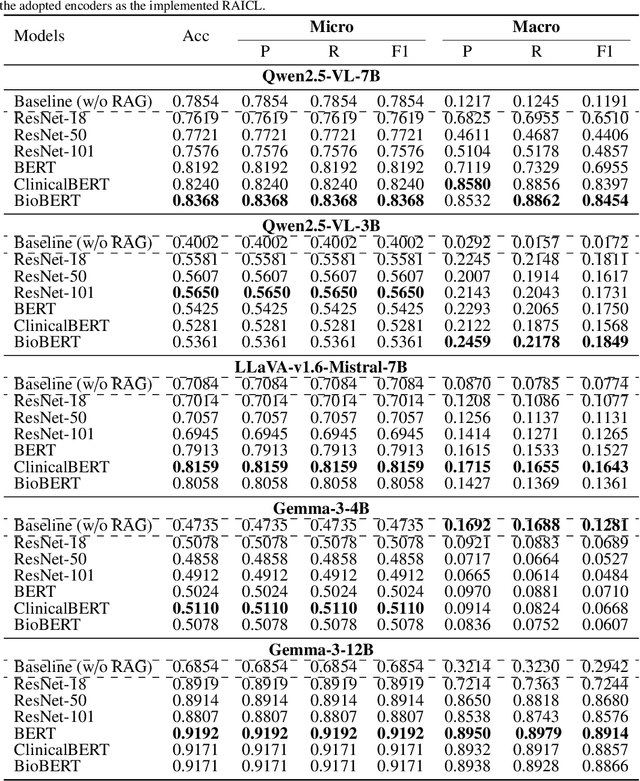
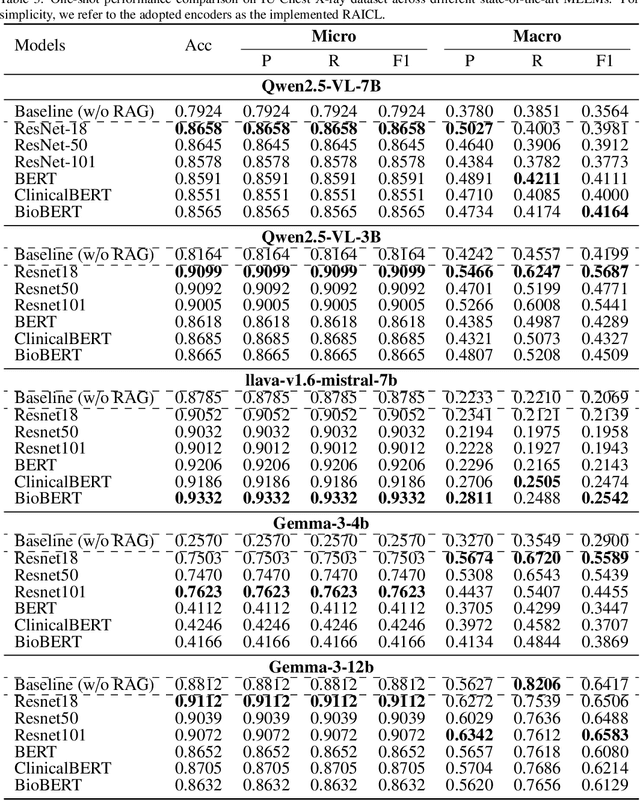
Abstract:Objectives: We aim to dynamically retrieve informative demonstrations, enhancing in-context learning in multimodal large language models (MLLMs) for disease classification. Methods: We propose a Retrieval-Augmented In-Context Learning (RAICL) framework, which integrates retrieval-augmented generation (RAG) and in-context learning (ICL) to adaptively select demonstrations with similar disease patterns, enabling more effective ICL in MLLMs. Specifically, RAICL examines embeddings from diverse encoders, including ResNet, BERT, BioBERT, and ClinicalBERT, to retrieve appropriate demonstrations, and constructs conversational prompts optimized for ICL. We evaluated the framework on two real-world multi-modal datasets (TCGA and IU Chest X-ray), assessing its performance across multiple MLLMs (Qwen, Llava, Gemma), embedding strategies, similarity metrics, and varying numbers of demonstrations. Results: RAICL consistently improved classification performance. Accuracy increased from 0.7854 to 0.8368 on TCGA and from 0.7924 to 0.8658 on IU Chest X-ray. Multi-modal inputs outperformed single-modal ones, with text-only inputs being stronger than images alone. The richness of information embedded in each modality will determine which embedding model can be used to get better results. Few-shot experiments showed that increasing the number of retrieved examples further enhanced performance. Across different similarity metrics, Euclidean distance achieved the highest accuracy while cosine similarity yielded better macro-F1 scores. RAICL demonstrated consistent improvements across various MLLMs, confirming its robustness and versatility. Conclusions: RAICL provides an efficient and scalable approach to enhance in-context learning in MLLMs for multimodal disease classification.
EPEE: Towards Efficient and Effective Foundation Models in Biomedicine
Mar 03, 2025Abstract:Foundation models, including language models, e.g., GPT, and vision models, e.g., CLIP, have significantly advanced numerous biomedical tasks. Despite these advancements, the high inference latency and the "overthinking" issues in model inference impair the efficiency and effectiveness of foundation models, thus limiting their application in real-time clinical settings. To address these challenges, we proposed EPEE (Entropy- and Patience-based Early Exiting), a novel hybrid strategy designed to improve the inference efficiency of foundation models. The core idea was to leverage the strengths of entropy-based and patience-based early exiting methods to overcome their respective weaknesses. To evaluate EPEE, we conducted experiments on three core biomedical tasks-classification, relation extraction, and event extraction-using four foundation models (BERT, ALBERT, GPT-2, and ViT) across twelve datasets, including clinical notes and medical images. The results showed that EPEE significantly reduced inference time while maintaining or improving accuracy, demonstrating its adaptability to diverse datasets and tasks. EPEE addressed critical barriers to deploying foundation models in healthcare by balancing efficiency and effectiveness. It potentially provided a practical solution for real-time clinical decision-making with foundation models, supporting reliable and efficient workflows.
MMRAG: Multi-Mode Retrieval-Augmented Generation with Large Language Models for Biomedical In-Context Learning
Feb 21, 2025



Abstract:Objective: To optimize in-context learning in biomedical natural language processing by improving example selection. Methods: We introduce a novel multi-mode retrieval-augmented generation (MMRAG) framework, which integrates four retrieval strategies: (1) Random Mode, selecting examples arbitrarily; (2) Top Mode, retrieving the most relevant examples based on similarity; (3) Diversity Mode, ensuring variation in selected examples; and (4) Class Mode, selecting category-representative examples. This study evaluates MMRAG on three core biomedical NLP tasks: Named Entity Recognition (NER), Relation Extraction (RE), and Text Classification (TC). The datasets used include BC2GM for gene and protein mention recognition (NER), DDI for drug-drug interaction extraction (RE), GIT for general biomedical information extraction (RE), and HealthAdvice for health-related text classification (TC). The framework is tested with two large language models (Llama2-7B, Llama3-8B) and three retrievers (Contriever, MedCPT, BGE-Large) to assess performance across different retrieval strategies. Results: The results from the Random mode indicate that providing more examples in the prompt improves the model's generation performance. Meanwhile, Top mode and Diversity mode significantly outperform Random mode on the RE (DDI) task, achieving an F1 score of 0.9669, a 26.4% improvement. Among the three retrievers tested, Contriever outperformed the other two in a greater number of experiments. Additionally, Llama 2 and Llama 3 demonstrated varying capabilities across different tasks, with Llama 3 showing a clear advantage in handling NER tasks. Conclusion: MMRAG effectively enhances biomedical in-context learning by refining example selection, mitigating data scarcity issues, and demonstrating superior adaptability for NLP-driven healthcare applications.
PaperHelper: Knowledge-Based LLM QA Paper Reading Assistant
Feb 20, 2025Abstract:In the paper, we introduce a paper reading assistant, PaperHelper, a potent tool designed to enhance the capabilities of researchers in efficiently browsing and understanding scientific literature. Utilizing the Retrieval-Augmented Generation (RAG) framework, PaperHelper effectively minimizes hallucinations commonly encountered in large language models (LLMs), optimizing the extraction of accurate, high-quality knowledge. The implementation of advanced technologies such as RAFT and RAG Fusion significantly boosts the performance, accuracy, and reliability of the LLMs-based literature review process. Additionally, PaperHelper features a user-friendly interface that facilitates the batch downloading of documents and uses the Mermaid format to illustrate structural relationships between documents. Experimental results demonstrate that PaperHelper, based on a fine-tuned GPT-4 API, achieves an F1 Score of 60.04, with a latency of only 5.8 seconds, outperforming the basic RAG model by 7\% in F1 Score.
Towards Better Multi-task Learning: A Framework for Optimizing Dataset Combinations in Large Language Models
Dec 16, 2024



Abstract:To efficiently select optimal dataset combinations for enhancing multi-task learning (MTL) performance in large language models, we proposed a novel framework that leverages a neural network to predict the best dataset combinations. The framework iteratively refines the selection, greatly improving efficiency, while being model-, dataset-, and domain-independent. Through experiments on 12 biomedical datasets across four tasks - named entity recognition, relation extraction, event extraction, and text classification-we demonstrate that our approach effectively identifies better combinations, even for tasks that may seem unpromising from a human perspective. This verifies that our framework provides a promising solution for maximizing MTL potential.
RAMIE: Retrieval-Augmented Multi-task Information Extraction with Large Language Models on Dietary Supplements
Nov 24, 2024Abstract:\textbf{Objective:} We aimed to develop an advanced multi-task large language model (LLM) framework to extract multiple types of information about dietary supplements (DS) from clinical records. \textbf{Methods:} We used four core DS information extraction tasks - namely, named entity recognition (NER: 2,949 clinical sentences), relation extraction (RE: 4,892 sentences), triple extraction (TE: 2,949 sentences), and usage classification (UC: 2,460 sentences) as our multitasks. We introduced a novel Retrieval-Augmented Multi-task Information Extraction (RAMIE) Framework, including: 1) employed instruction fine-tuning techniques with task-specific prompts, 2) trained LLMs for multiple tasks with improved storage efficiency and lower training costs, and 3) incorporated retrieval augmentation generation (RAG) techniques by retrieving similar examples from the training set. We compared RAMIE's performance to LLMs with instruction fine-tuning alone and conducted an ablation study to assess the contributions of multi-task learning and RAG to improved multitasking performance. \textbf{Results:} With the aid of the RAMIE framework, Llama2-13B achieved an F1 score of 87.39 (3.51\% improvement) on the NER task and demonstrated outstanding performance on the RE task with an F1 score of 93.74 (1.15\% improvement). For the TE task, Llama2-7B scored 79.45 (14.26\% improvement), and MedAlpaca-7B achieved the highest F1 score of 93.45 (0.94\% improvement) on the UC task. The ablation study revealed that while MTL increased efficiency with a slight trade-off in performance, RAG significantly boosted overall accuracy. \textbf{Conclusion:} This study presents a novel RAMIE framework that demonstrates substantial improvements in multi-task information extraction for DS-related data from clinical records. Our framework can potentially be applied to other domains.
Large Language Models for Disease Diagnosis: A Scoping Review
Aug 27, 2024Abstract:Automatic disease diagnosis has become increasingly valuable in clinical practice. The advent of large language models (LLMs) has catalyzed a paradigm shift in artificial intelligence, with growing evidence supporting the efficacy of LLMs in diagnostic tasks. Despite the growing attention in this field, many critical research questions remain under-explored. For instance, what diseases and LLM techniques have been investigated for diagnostic tasks? How can suitable LLM techniques and evaluation methods be selected for clinical decision-making? To answer these questions, we performed a comprehensive analysis of LLM-based methods for disease diagnosis. This scoping review examined the types of diseases, associated organ systems, relevant clinical data, LLM techniques, and evaluation methods reported in existing studies. Furthermore, we offered guidelines for data preprocessing and the selection of appropriate LLM techniques and evaluation strategies for diagnostic tasks. We also assessed the limitations of current research and delineated the challenges and future directions in this research field. In summary, our review outlined a blueprint for LLM-based disease diagnosis, helping to streamline and guide future research endeavors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge