Yongkang Xiao
Retrieval-augmented in-context learning for multimodal large language models in disease classification
May 04, 2025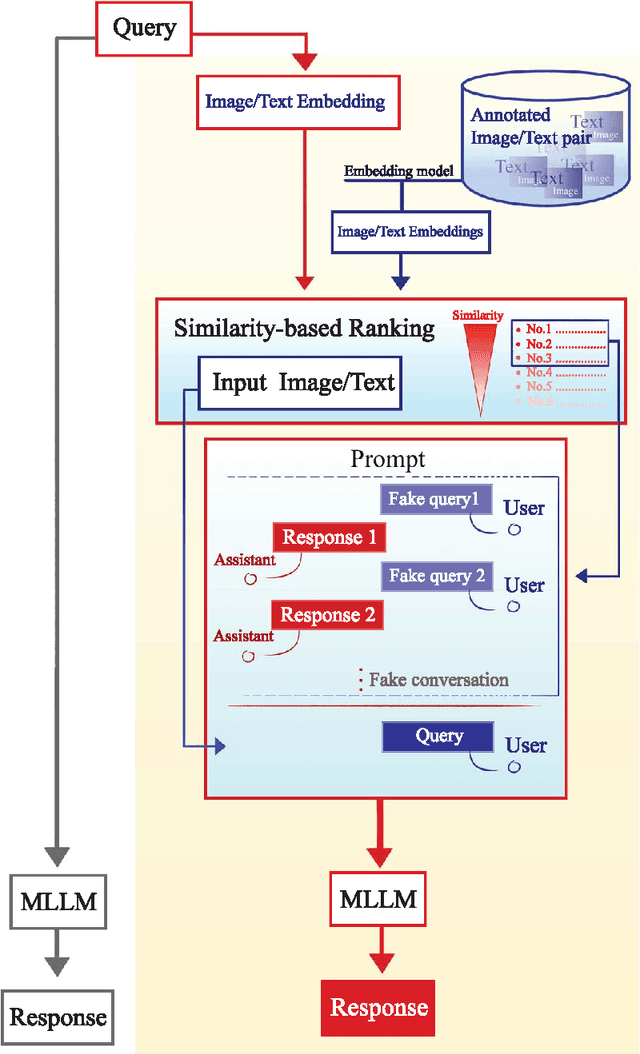
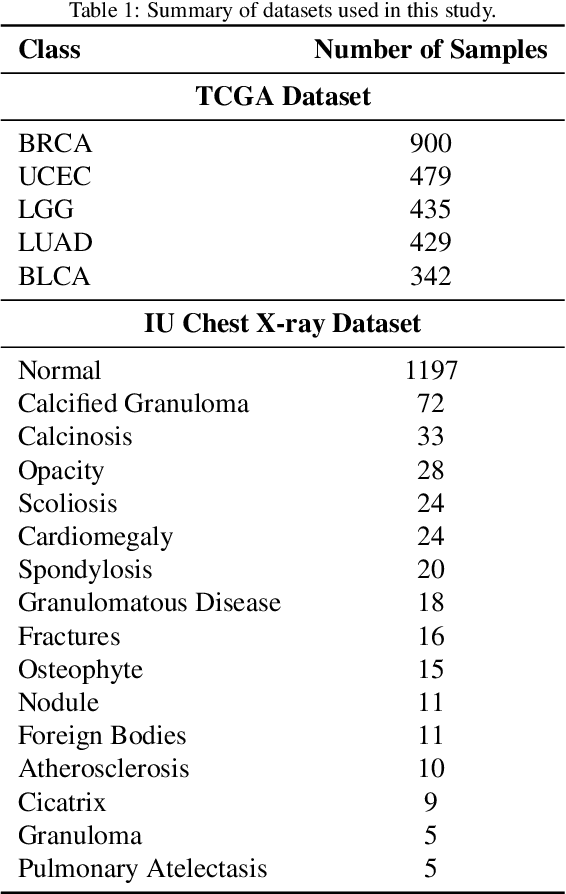
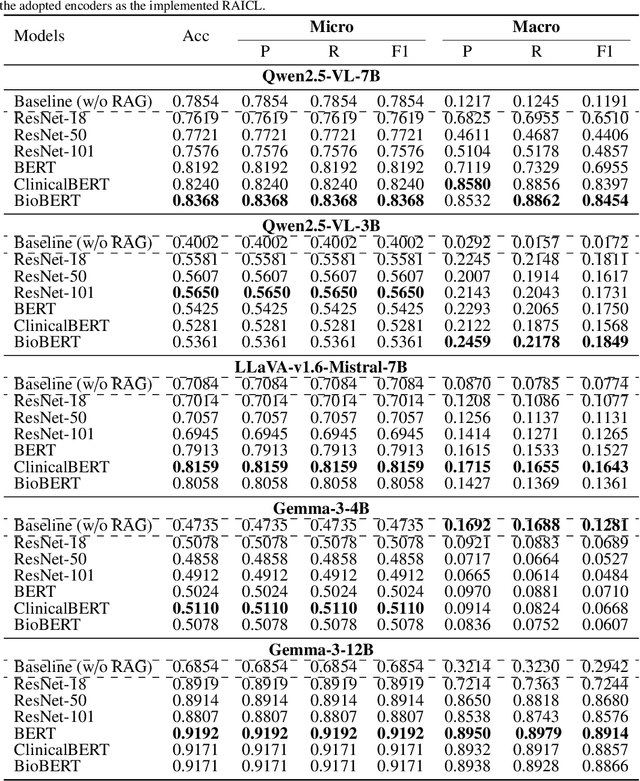
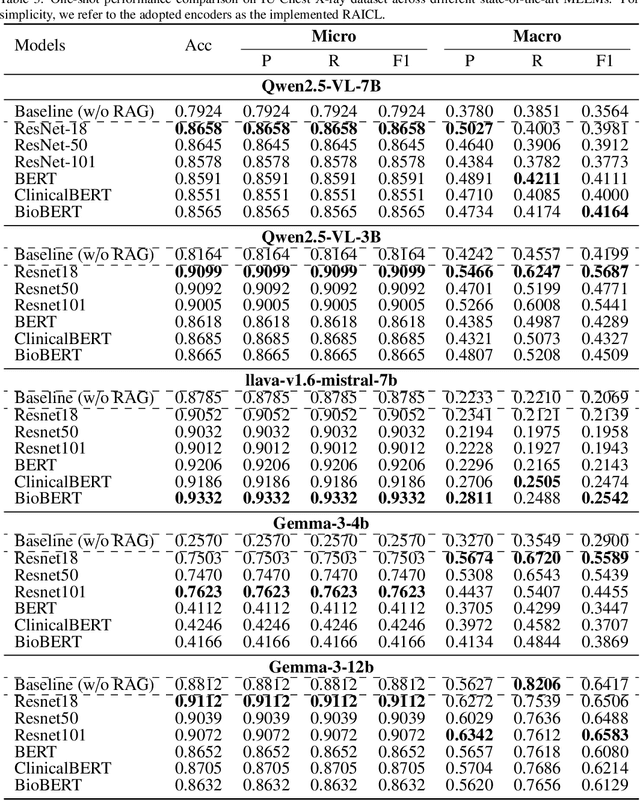
Abstract:Objectives: We aim to dynamically retrieve informative demonstrations, enhancing in-context learning in multimodal large language models (MLLMs) for disease classification. Methods: We propose a Retrieval-Augmented In-Context Learning (RAICL) framework, which integrates retrieval-augmented generation (RAG) and in-context learning (ICL) to adaptively select demonstrations with similar disease patterns, enabling more effective ICL in MLLMs. Specifically, RAICL examines embeddings from diverse encoders, including ResNet, BERT, BioBERT, and ClinicalBERT, to retrieve appropriate demonstrations, and constructs conversational prompts optimized for ICL. We evaluated the framework on two real-world multi-modal datasets (TCGA and IU Chest X-ray), assessing its performance across multiple MLLMs (Qwen, Llava, Gemma), embedding strategies, similarity metrics, and varying numbers of demonstrations. Results: RAICL consistently improved classification performance. Accuracy increased from 0.7854 to 0.8368 on TCGA and from 0.7924 to 0.8658 on IU Chest X-ray. Multi-modal inputs outperformed single-modal ones, with text-only inputs being stronger than images alone. The richness of information embedded in each modality will determine which embedding model can be used to get better results. Few-shot experiments showed that increasing the number of retrieved examples further enhanced performance. Across different similarity metrics, Euclidean distance achieved the highest accuracy while cosine similarity yielded better macro-F1 scores. RAICL demonstrated consistent improvements across various MLLMs, confirming its robustness and versatility. Conclusions: RAICL provides an efficient and scalable approach to enhance in-context learning in MLLMs for multimodal disease classification.
The Efficiency vs. Accuracy Trade-off: Optimizing RAG-Enhanced LLM Recommender Systems Using Multi-Head Early Exit
Jan 04, 2025



Abstract:The deployment of Large Language Models (LLMs) in recommender systems for predicting Click-Through Rates (CTR) necessitates a delicate balance between computational efficiency and predictive accuracy. This paper presents an optimization framework that combines Retrieval-Augmented Generation (RAG) with an innovative multi-head early exit architecture to concurrently enhance both aspects. By integrating Graph Convolutional Networks (GCNs) as efficient retrieval mechanisms, we are able to significantly reduce data retrieval times while maintaining high model performance. The early exit strategy employed allows for dynamic termination of model inference, utilizing real-time predictive confidence assessments across multiple heads. This not only quickens the responsiveness of LLMs but also upholds or improves their accuracy, making it ideal for real-time application scenarios. Our experiments demonstrate how this architecture effectively decreases computation time without sacrificing the accuracy needed for reliable recommendation delivery, establishing a new standard for efficient, real-time LLM deployment in commercial systems.
RiTeK: A Dataset for Large Language Models Complex Reasoning over Textual Knowledge Graphs
Oct 17, 2024



Abstract:Answering complex real-world questions often requires accurate retrieval from textual knowledge graphs (TKGs). The scarcity of annotated data, along with intricate topological structures, makes this task particularly challenging. As the nature of relational path information could enhance the inference ability of Large Language Models (LLMs), efficiently retrieving more complex relational path information from TKGs presents another key challenge. To tackle these challenges, we first develop a Dataset for LLMs Complex Reasoning over Textual Knowledge Graphs (RiTeK) with a broad topological structure coverage.We synthesize realistic user queries that integrate diverse topological structures, relational information, and complex textual descriptions. We conduct rigorous expert evaluation to validate the quality of our synthesized queries. And then, we introduce an enhanced Monte Carlo Tree Search (MCTS) method, Relational MCTS, to automatically extract relational path information from textual graphs for specific queries. Our dataset mainly covers the medical domain as the relation types and entity are complex and publicly available. Experimental results indicate that RiTeK poses significant challenges for current retrieval and LLM systems, while the proposed Relational MCTS method enhances LLM inference ability and achieves state-of-the-art performance on RiTeK.
Benchmarking Retrieval-Augmented Large Language Models in Biomedical NLP: Application, Robustness, and Self-Awareness
May 16, 2024Abstract:Large language models (LLM) have demonstrated remarkable capabilities in various biomedical natural language processing (NLP) tasks, leveraging the demonstration within the input context to adapt to new tasks. However, LLM is sensitive to the selection of demonstrations. To address the hallucination issue inherent in LLM, retrieval-augmented LLM (RAL) offers a solution by retrieving pertinent information from an established database. Nonetheless, existing research work lacks rigorous evaluation of the impact of retrieval-augmented large language models on different biomedical NLP tasks. This deficiency makes it challenging to ascertain the capabilities of RAL within the biomedical domain. Moreover, the outputs from RAL are affected by retrieving the unlabeled, counterfactual, or diverse knowledge that is not well studied in the biomedical domain. However, such knowledge is common in the real world. Finally, exploring the self-awareness ability is also crucial for the RAL system. So, in this paper, we systematically investigate the impact of RALs on 5 different biomedical tasks (triple extraction, link prediction, classification, question answering, and natural language inference). We analyze the performance of RALs in four fundamental abilities, including unlabeled robustness, counterfactual robustness, diverse robustness, and negative awareness. To this end, we proposed an evaluation framework to assess the RALs' performance on different biomedical NLP tasks and establish four different testbeds based on the aforementioned fundamental abilities. Then, we evaluate 3 representative LLMs with 3 different retrievers on 5 tasks over 9 datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge